Comparative analysis of protein domain organization
- PMID: 14993202
- PMCID: PMC535408
- DOI: 10.1101/gr.1610504
Comparative analysis of protein domain organization
Abstract
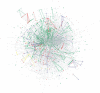
We have developed a set of graph theory-based tools, which we call Comparative Analysis of Protein Domain Organization (CADO), to survey and compare protein domain organizations of different organisms. In the language of CADO, the organization of protein domains in a given organism is shown as a domain graph in which protein domains are represented as vertices, and domain combinations, defined as instances of two domains found in one protein, are represented as edges. CADO provides a new way to analyze and compare whole proteomes, including identifying the consensus and difference of domain organization between organisms. CADO was used to analyze and compare >50 bacterial, archaeal, and eukaryotic genomes. Examples and overviews presented here include the analysis of the modularity of domain graphs and the functional study of domains based on the graph topology. We also report on the results of comparing domain graphs of two organisms, Pyrococcus horikoshii (an extremophile) and Haemophilus influenzae (a parasite with reduced genome) with other organisms. Our comparison provides new insights into the genome organization of these organisms. Finally, we report on the specific domain combinations characterizing the three kingdoms of life, and the kingdom "signature" domain organizations derived from those specific domain combinations.
Figures




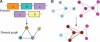
References
-
- Aasland, R., Gibson, T.J., and Stewart, A.F. 1995. The PHD finger: Implications for chromatin-mediated transcriptional regulation. Trends Biochem. Sci. 20: 56–59. - PubMed
-
- Anantharaman, V., Koonin, E.V., and Aravind, L. 2001. TRAM, a predicted RNA-binding domain, common to tRNA uracil methylation and adenine thiolation enzymes. FEMS Microbiol. Lett. 197: 215–221. - PubMed
-
- Apic, G., Gough, J., and Teichmann, S.A. 2001. Domain combinations in archaeal, eubacterial and eukaryotic proteomes. J. Mol. Biol. 310: 311–325. - PubMed
-
- Aravind, L. and Koonin, E.V. 1998. The HD domain defines a new superfamily of metal-dependent phosphohydrolases. Trends Biochem. Sci. 23: 469–472. - PubMed
-
- Aravind, L. and Koonin, E.V. 2000. The U box is a modified RING finger—A common domain in ubiquitination. Curr. Biol. 10: R132–R134. - PubMed
WEB SITE REFERENCES
-
- ftp://ftp.ncbi.nih.gov/; NCBI GenBank.
-
- http://genome.jgi-psf.org/ciona4/ciona4.download.ftp.html; Ciona intestinalis.
-
- http://genome.jgi-psf.org/fugu6/fugu6.download.ftp.html; Fugu rubripes sequence.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
