Gene conversion and the evolution of protocadherin gene cluster diversity
- PMID: 14993203
- PMCID: PMC353213
- DOI: 10.1101/gr.2133704
Gene conversion and the evolution of protocadherin gene cluster diversity
Abstract
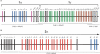
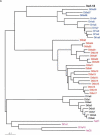
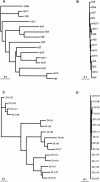
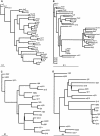
The synaptic cell adhesion molecules encoded by the protocadherin gene cluster are hypothesized to provide a molecular code involved in the generation of synaptic complexity in the developing brain. Variation in copy number and sequence content of protocadherin cluster genes among vertebrate species could reflect adaptive differences in protocadherin function. We have completed an analysis of zebrafish protocadherin cluster genes. Zebrafish have two unlinked protocadherin clusters, DrPcdh1 and DrPcdh2. Like mammalian protocadherin clusters, DrPcdh1 has both alpha and gamma variable and constant region exons. A consensus protocadherin promoter motif sequence identified in mammals is also conserved in zebrafish. Few orthologous relationships, however, are apparent between zebrafish and mammalian protocadherin proteins. Here we show that protocadherin cluster genes in human, mouse, rat, and zebrafish are subject to striking gene conversion events. These events are restricted to regions of the coding sequence, particularly the coding sequences of ectodomain 6 and the cytoplasmic domain. Diversity among paralogs is restricted to particular ectodomains that are excluded from conversion events. Conversion events are also strongly correlated with an increase in third-position GC content. We propose that the combination of lineage-specific duplication, restricted gene conversion, and adaptive variation in diversified ectodomains drives vertebrate protocadherin cluster evolution.
Figures



References
-
- Amores, A., Force, A., Yan, Y.L., Joly, L., Amemiya, C., Fritz, A., Ho, R.K., Langeland, J., Prince, V., Wang, Y.L., et al. 1998. Zebrafish hox clusters and vertebrate genome evolution. Science 282: 1711–1714. - PubMed
-
- Bailey, T. and Elkan, C. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology, pp. 28–36, AAAI Press, Menlo Park, CA. - PubMed
-
- Birdsell, J.A. 2002. Integrating genomics, bioinformatics, and classical genetics to study the effects of recombination on genome evolution. Mol. Biol. Evol. 19: 1181–1197. - PubMed
WEB SITE REFERENCES
-
- http://hmmer.wustl.edu/; HMMER.
-
- http://www.math.wustl.edu/~sawyer/geneconv; GeneConv.
-
- http://www.cbs.dtu.dk/services/RevTrans/; RevTrans.
-
- http://genome.ucsc.edu/; UCSC Genome Browser.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
