Mapping of a functional recombination motif that defines isotype specificity for mu-->gamma3 switch recombination implicates NF-kappaB p50 as the isotype-specific switching factor
- PMID: 14993249
- PMCID: PMC2213297
- DOI: 10.1084/jem.20031935
Mapping of a functional recombination motif that defines isotype specificity for mu-->gamma3 switch recombination implicates NF-kappaB p50 as the isotype-specific switching factor
Abstract
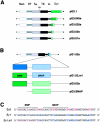
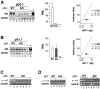
Ig class switch recombination (CSR) requires expression of activation-induced deaminase (AID) and production of germline transcripts to target S regions for recombination. However, the mechanism of CSR remains unclear. Here we show that an extrachromosomal S plasmid assay is AID dependent and that a single consensus repeat is both necessary and sufficient for isotype-specific CSR. Transfected switch substrates specific for mu-->gamma3 and mu-->gamma1 are stimulated to switch with lipopolysaccharide (LPS) alone or LPS and interleukin-4, respectively. An Sgamma3/Sgamma1 substrate containing only three Sgamma3-associated nucleotides reconstituted LPS responsiveness and permitted mapping of a functional recombination motif specific for mu-->gamma3 CSR. This functional recombination motif colocalized with a binding site for NF-kappaB p50, and p50 binding to this site was previously established. We show a p50 requirement for plasmid-based mu-->gamma3 CSR using p50-deficient B cells. Switch junctions from p50-deficient B cells showed decreased lengths of microhomology between Smu and Sgamma3 relative to wild-type cells, indicating a function for p50 in the mechanics of CSR. We note a striking parallel between the affects of p50 and Msh2 deficiency on Smu/Sgamma3 junctions. The data suggest that p50 may be the isotype-specific factor in mu-->gamma3 CSR and epistatic with Msh2.
Figures



References
-
- Stavnezer, J. 2000. Molecular processes that regulate class switching. Curr. Top. Microbiol. Immunol. 245:127–168. - PubMed
-
- Manis, J.P., M. Tian, and F.W. Alt. 2002. Mechanism and control of class-switch recombination. Trends Immunol. 23:31–39. - PubMed
-
- Muramatsu, M., K. Kinoshita, S. Fagarasan, S. Yamada, Y. Shinkai, and T. Honjo. 2000. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 102:553–563. - PubMed
-
- Muramatsu, M., V.S. Sankaranand, S. Anant, M. Sugai, K. Kinoshita, N.O. Davidson, and T. Honjo. 1999. Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J. Biol. Chem. 274:18470–18476. - PubMed
-
- Revy, P., T. Muto, Y. Levy, F. Geissmann, A. Plebani, O. Sanal, N. Catalan, M. Forveille, R. Dufourcq-Labelouse, A. Gennery, et al. 2000. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2). Cell. 102:565–575. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

