A novel virulence gene in Klebsiella pneumoniae strains causing primary liver abscess and septic metastatic complications
- PMID: 14993253
- PMCID: PMC2213305
- DOI: 10.1084/jem.20030857
A novel virulence gene in Klebsiella pneumoniae strains causing primary liver abscess and septic metastatic complications
Abstract
Primary Klebsiella pneumoniae liver abscess complicated with metastatic meningitis or endophthalmitis is a globally emerging infectious disease. Its pathogenic mechanism remains unclear. The bacterial virulence factors were explored by comparing clinical isolates. Differences in mucoviscosity were observed between strains that caused primary liver abscess (invasive) and those that did not (noninvasive). Hypermucoviscosity correlated with a high serum resistance and was more prevalent in invasive strains (52/53 vs. 9/52; P < 0.0001). Transposon mutagenesis identified candidate virulence genes. A novel 1.2-kb locus, magA, which encoded a 43-kD outer membrane protein, was significantly more prevalent in invasive strains (52/53 vs. 14/52; P < 0.0001). The wild-type strain produced a mucoviscous exopolysaccharide web, actively proliferated in nonimmune human serum, resisted phagocytosis, and caused liver microabscess and meningitis in mice. However, magA- mutants lost the exopolysaccharide web and became extremely serum sensitive, phagocytosis susceptible, and avirulent to mice. Virulence was restored by complementation using a magA-containing plasmid. We conclude that magA fits molecular Koch's postulates as a virulence gene. Thus, this locus can be used as a marker for the rapid diagnosis and for tracing the source of this emerging infectious disease.
Figures

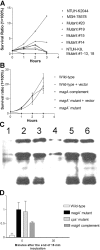

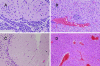


References
-
- Abbott, S. 1999. Klebsiella, Enterobacter, Citrobacter and Serratia. Manual of Clinical Microbiology. 7th ed. P.R. Murray, E.J. Baron, M.A. Pfaller, F.C. Tenover, and R.H. Yolken, editors. American Society for Microbiology Press, Washington D.C. 475–482.
-
- Eisenstein, B.I., and D.F. Zaleznik. 2000. Enterobacteriaceae. Principles and Practice of Infectious Diseases. 5th ed. G.L. Mandell, J.E. Bennett, and R. Dolin, editors. Churchill-Livingstone, Philadelphia. 2294–2309.
-
- Wang, J.H., Y.C. Liu, S.S. Lee, M.Y. Yen, Y.S. Chen, J.H. Wang, S.R. Wann, and H.H. Lin. 1998. Primary liver abscess due to Klebsiella pneumoniae in Taiwan. Clin. Infect. Dis. 26:1434–1438. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

