Replication stalling at Friedreich's ataxia (GAA)n repeats in vivo
- PMID: 14993268
- PMCID: PMC355872
- DOI: 10.1128/MCB.24.6.2286-2295.2004
Replication stalling at Friedreich's ataxia (GAA)n repeats in vivo
Abstract
Friedreich's ataxia (GAA)n repeats of various lengths were cloned into a Saccharymyces cerevisiae plasmid, and their effects on DNA replication were analyzed using two-dimensional electrophoresis of replication intermediates. We found that premutation- and disease-size repeats stalled the replication fork progression in vivo, while normal-size repeats did not affect replication. Remarkably, the observed threshold repeat length for replication stalling in yeast (approximately 40 repeats) closely matched the threshold length for repeat expansion in humans. Further, replication stalling was strikingly orientation dependent, being pronounced only when the repeat's homopurine strand served as the lagging strand template. Finally, it appeared that length polymorphism of the (GAA)n. (TTC)n repeat in both expansions and contractions drastically increases in the repeat's orientation that is responsible for the replication stalling. These data represent the first direct proof of the effects of (GAA)n repeats on DNA replication in vivo. We believe that repeat-caused replication attenuation in vivo is due to triplex formation. The apparent link between the replication stalling and length polymorphism of the repeat points to a new model for the repeat expansion.
Figures

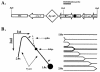




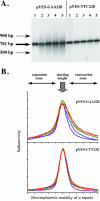
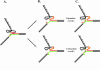
References
-
- Bowater, R. P., and R. D. Wells. 2001. The intrinsically unstable life of DNA triplet repeats associated with human hereditary disorders. Prog. Nucleic Acid Res. Mol. Biol. 66:159-202. - PubMed
-
- Brewer, B. J., and W. L. Fangman. 1987. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell 51:463-471. - PubMed
-
- Campuzano, V., L. Montermini, Y. Lutz, L. Cova, C. Hindelag, S. Jiralerspong, Y. Trottier, S. J. Kish, B. Faucheux, P. Trouillas, F. J. Authier, A. Durr, J.-L. Mandel, A. L. Vescovi, M. Pandolfo, and M. Koenig. 1997. Frataxin is reduced in Friedreich ataxia patients and is associated with mitochondrial membranes. Hum. Mol. Genet. 6:1771-1780. - PubMed
-
- Campuzano, V., L. Montermini, M. D. Molto, L. Pianese, M. Cossee, F. Cavalcanti, E. Monros, F. Rodius, F. Duclos, A. Monticelli, F. Zara, J. Canizares, H. Koutnikova, S. I. Bidichandari, C. Gellera, A. Brice, P. Trouillas, G. De Michele, A. Filla, R. De Frutos, F. Palau, P. I. Patel, S. Di Donato, J.-L. Mandel, S. Cocozza, M. Koenig, and M. Pandolfo. 1996. Friedreich's ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science 271:1423-1427. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
