Elongation inhibition by DRB sensitivity-inducing factor is regulated by the A20 promoter via a novel negative element and NF-kappaB
- PMID: 14993282
- PMCID: PMC355833
- DOI: 10.1128/MCB.24.6.2444-2454.2004
Elongation inhibition by DRB sensitivity-inducing factor is regulated by the A20 promoter via a novel negative element and NF-kappaB
Abstract
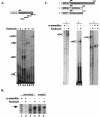
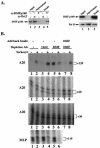
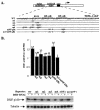
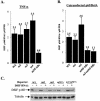
A20 is an immediate-early NF-kappaB target gene. Prior to NF-kappaB stimulation, the A20 promoter is bound by the polymerase II machinery to allow rapid transcription activation. Here we show that the basal A20 transcription is repressed at the level of elongation in a promoter-specific fashion. Immunodepletion in vitro and RNA interference in cultured cells suggest that the basal elongation inhibition is conferred by DRB sensitivity-inducing factor (DSIF). We have identified a negative upstream promoter element called ELIE that controls DSIF activity. Remarkably, following NF-kappaB stimulation, inhibition of the A20 promoter by DSIF persists, but it is now regulated by NF-kappaB rather than ELIE. Similar regulation by DSIF is shown for another NF-kappaB-responsive gene, the IkappaBalpha gene. These findings reveal an intimate and dynamic relationship between DSIF inhibition of elongation and promoter-bound transcription factors. The potential significance of the differential regulation of DSIF activity by cis-acting elements is discussed.
Figures




Similar articles
-
Interplay between E-box and NF-kappaB in regulation of A20 gene by DRB sensitivity-inducing factor (DSIF).J Biol Chem. 2008 Jan 18;283(3):1317-1323. doi: 10.1074/jbc.M706767200. Epub 2007 Oct 25. J Biol Chem. 2008. PMID: 17962196
-
DSIF restricts NF-κB signaling by coordinating elongation with mRNA processing of negative feedback genes.Cell Rep. 2012 Oct 25;2(4):722-31. doi: 10.1016/j.celrep.2012.08.041. Epub 2012 Oct 4. Cell Rep. 2012. PMID: 23041311
-
The RING ubiquitin E3 RNF114 interacts with A20 and modulates NF-κB activity and T-cell activation.Cell Death Dis. 2014 Aug 28;5(8):e1399. doi: 10.1038/cddis.2014.366. Cell Death Dis. 2014. PMID: 25165885 Free PMC article.
-
ABINs: A20 binding inhibitors of NF-kappa B and apoptosis signaling.Biochem Pharmacol. 2009 Jul 15;78(2):105-14. doi: 10.1016/j.bcp.2009.02.009. Epub 2009 Feb 27. Biochem Pharmacol. 2009. PMID: 19464428 Review.
-
Transcription elongation factors DSIF and NELF: promoter-proximal pausing and beyond.Biochim Biophys Acta. 2013 Jan;1829(1):98-104. doi: 10.1016/j.bbagrm.2012.11.007. Epub 2012 Nov 29. Biochim Biophys Acta. 2013. PMID: 23202475 Review.
Cited by
-
An enhancer element harboring variants associated with systemic lupus erythematosus engages the TNFAIP3 promoter to influence A20 expression.PLoS Genet. 2013;9(9):e1003750. doi: 10.1371/journal.pgen.1003750. Epub 2013 Sep 5. PLoS Genet. 2013. PMID: 24039598 Free PMC article.
-
Promoter proximal pausing on genes in metazoans.Chromosoma. 2009 Feb;118(1):1-10. doi: 10.1007/s00412-008-0182-4. Epub 2008 Oct 2. Chromosoma. 2009. PMID: 18830703 Review.
-
TAF4/4b x TAF12 displays a unique mode of DNA binding and is required for core promoter function of a subset of genes.J Biol Chem. 2009 Sep 25;284(39):26286-96. doi: 10.1074/jbc.M109.011486. Epub 2009 Jul 27. J Biol Chem. 2009. PMID: 19635797 Free PMC article.
-
Attenuation of estrogen receptor alpha-mediated transcription through estrogen-stimulated recruitment of a negative elongation factor.Genes Dev. 2004 Sep 1;18(17):2134-46. doi: 10.1101/gad.1214104. Genes Dev. 2004. PMID: 15342491 Free PMC article.
-
Role of A20 in cIAP-2 protection against tumor necrosis factor α (TNF-α)-mediated apoptosis in endothelial cells.Int J Mol Sci. 2014 Mar 3;15(3):3816-33. doi: 10.3390/ijms15033816. Int J Mol Sci. 2014. PMID: 24595242 Free PMC article.
References
-
- Barboric, M., R. M. Nissen, S. Kanazawa, N. Jabrane-Ferrat, and B. M. Peterlin. 2001. NF-κB binds P-TEFb to stimulate transcriptional elongation by RNA polymerase II. Mol. Cell 8:327-337. - PubMed
-
- Brummelkamp, T. R., R. Bernards, and R. Agami. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296:550-555. - PubMed
-
- Conaway, J. W., A. Shilatifard, A. Dvir, and R. C. Conaway. 2000. Control of elongation by RNA polymerase II. Trends Biochem. Sci. 25:375-380. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
