Histone H3-K9 methyltransferase ESET is essential for early development
- PMID: 14993285
- PMCID: PMC355869
- DOI: 10.1128/MCB.24.6.2478-2486.2004
Histone H3-K9 methyltransferase ESET is essential for early development
Abstract
Methylation of histone H3 at lysine 9 (H3-K9) mediates heterochromatin formation by forming a binding site for HP1 and also participates in silencing gene expression at euchromatic sites. ESET, G9a, SUV39-h1, SUV39-h2, and Eu-HMTase are histone methyltransferases that catalyze H3-K9 methylation in mammalian cells. Previous studies demonstrate that the SUV39-h proteins are preferentially targeted to the pericentric heterochromatin, and mice lacking both Suv39-h genes show cytogenetic abnormalities and an increased incidence of lymphoma. G9a methylates H3-K9 in euchromatin, and G9a null embryos die at 8.5 days postcoitum (dpc). G9a null embryo stem (ES) cells show altered DNA methylation in the Prader-Willi imprinted region and ectopic expression of the Mage genes. So far, an Eu-HMTase mouse knockout has not been reported. ESET catalyzes methylation of H3-K9 and localizes mainly in euchromatin. To investigate the in vivo function of Eset, we have generated an allele that lacks the entire pre- and post-SET domains and that expresses lacZ under the endogenous regulation of the Eset gene. We found that zygotic Eset expression begins at the blastocyst stage and is ubiquitous during postimplantation mouse development, while the maternal Eset transcripts are present in oocytes and persist throughout preimplantation development. The homozygous mutations of Eset resulted in peri-implantation lethality between 3.5 and 5.5 dpc. Blastocysts null for Eset were recovered but in less than Mendelian ratios. Upon culturing, 18 of 24 Eset(-/-) blastocysts showed defective growth of the inner cell mass and, in contrast to the approximately 65% recovery of wild-type and Eset(+/-) ES cells, no Eset(-/-) ES cell lines were obtained. Global H3-K9 trimethylation and DNA methylation at IAP repeats in Eset(-/-) blastocyst outgrowths were not dramatically altered. Together, these results suggest that Eset is required for peri-implantation development and the survival of ES cells.
Figures
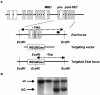


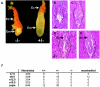

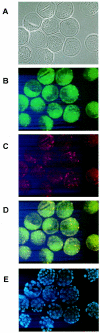
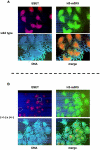
References
-
- Beard, C., E. Li, and R. Jaenisch. 1995. Loss of methylation activates Xist in somatic but not in embryonic cells. Genes Dev. 9:2325-2334. - PubMed
-
- Bourc'his, D., G. L. Xu, C. S. Lin, B. Bollman, and T. H. Bestor. 2001. Dnmt3L and the establishment of maternal genomic imprints. Science 294:2536-2539. - PubMed
-
- Dodge, J. E., A. F. List, and B. W. Futscher. 1998. Selective variegated methylation of the p15 CpG island in acute myeloid leukemia. Int. J. Cancer 78:561-567. - PubMed
-
- Gaudet, F., J. G. Hodgson, A. Eden, L. Jackson-Grusby, J. Dausman, J. W. Gray, H. Leonhardt, and R. Jaenisch. 2003. Induction of tumors in mice by genomic hypomethylation. Science 300:489-492. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
