Np95 is a histone-binding protein endowed with ubiquitin ligase activity
- PMID: 14993289
- PMCID: PMC355858
- DOI: 10.1128/MCB.24.6.2526-2535.2004
Np95 is a histone-binding protein endowed with ubiquitin ligase activity
Abstract
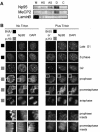
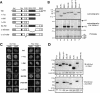
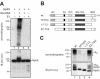
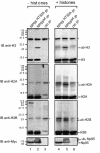
Np95 is an important determinant in cell cycle progression. Its expression is tightly regulated and becomes detectable shortly before the entry of cells into S phase. Accordingly, Np95 is absolutely required for the G1/S transition. Its continued expression throughout the S/G2/M phases further suggests additional roles. Indeed, Np95 has been implicated in DNA damage response. Here, we show that Np95 is tightly bound to chromatin in vivo and that it binds to histones in vivo and in vitro. The binding to histones is direct and shows a remarkable preference for histone H3 and its N-terminal tail. A novel protein domain, the SRA-YDG domain, contained in Np95 is indispensable both for the interaction with histones and for chromatin binding in vivo. Np95 contains a RING finger. We show that this domain confers E3 ubiquitin ligase activity on Np95, which is specific for core histones, in vitro. Finally, Np95 shows specific E3 activity for histone H3 when the endogenous core octamer, coimmunoprecipitating with Np95, is used as a substrate. Histone ubiquitination is an important determinant in the regulation of chromatin structure and gene transcription. Thus, the demonstration that Np95 is a chromatin-associated ubiquitin ligase suggests possible molecular mechanisms for its action as a cell cycle regulator.
Figures



References
-
- Baarends, W. M., J. W. Hoogerbrugge, H. P. Roest, M. Ooms, J. Vreeburg, J. H. Hoeijmakers, and J. A. Grootegoed. 1999. Histone ubiquitination and chromatin remodeling in mouse spermatogenesis. Dev. Biol. 207:322-333. - PubMed
-
- Baumbusch, L. O., T. Thorstensen, V. Krauss, A. Fischer, K. Naumann, R. Assalkhou, I. Schulz, G. Reuter, and R. B. Aalen. 2001. The Arabidopsis thaliana genome contains at least 29 active genes encoding SET domain proteins that can be assigned to four evolutionarily conserved classes. Nucleic Acids Res. 29:4319-4333. - PMC - PubMed
-
- Briggs, S. D., T. Xiao, Z. W. Sun, J. A. Caldwell, J. Shabanowitz, D. F. Hunt, C. D. Allis, and B. D. Strahl. 2002. Gene silencing: trans-histone regulatory pathway in chromatin. Nature 418:498. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
