Engineered single-chain, antiparallel, coiled coil mimics the MerR metal binding site
- PMID: 14996817
- PMCID: PMC355954
- DOI: 10.1128/JB.186.6.1861-1868.2004
Engineered single-chain, antiparallel, coiled coil mimics the MerR metal binding site
Abstract
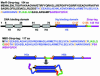
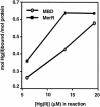
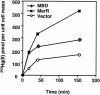
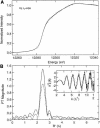
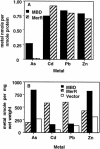
The repressor-activator MerR that controls transcription of the mercury resistance (mer) operon is unusual for its high sensitivity and specificity for Hg(II) in in vivo and in vitro transcriptional assays. The metal-recognition domain of MerR resides at the homodimer interface in a novel antiparallel arrangement of alpha-helix 5 that forms a coiled-coil motif. To facilitate the study of this novel metal binding motif, we assembled this antiparallel coiled coil into a single chain by directly fusing two copies of the 48-residue alpha-helix 5 of MerR. The resulting 107-residue polypeptide, called the metal binding domain (MBD), and wild-type MerR were overproduced and purified, and their metal-binding properties were determined in vivo and in vitro. In vitro MBD bound ca. 1.0 equivalent of Hg(II) per pair of binding sites, just as MerR does, and it showed only a slightly lower affinity for Hg(II) than did MerR. Extended X-ray absorption fine structure data showed that MBD has essentially the same Hg(II) coordination environment as MerR. In vivo, cells overexpressing MBD accumulated 70 to 100% more (203)Hg(II) than cells bearing the vector alone, without deleterious effects on cell growth. Both MerR and MBD variously bound other thiophilic metal ions, including Cd(II), Zn(II), Pb(II), and As(III), in vitro and in vivo. We conclude that (i) it is possible to simulate in a single polypeptide chain the in vitro and in vivo metal-binding ability of dimeric, full-length MerR and (ii) MerR's specificity in transcriptional activation does not reside solely in the metal-binding step.
Figures
Similar articles
-
Cd(II)-responsive and constitutive mutants implicate a novel domain in MerR.J Bacteriol. 1999 Jun;181(11):3462-71. doi: 10.1128/JB.181.11.3462-3471.1999. J Bacteriol. 1999. PMID: 10348859 Free PMC article.
-
Hg(II) sequestration and protection by the MerR metal-binding domain (MBD).Microbiology (Reading). 2006 Mar;152(Pt 3):709-719. doi: 10.1099/mic.0.28474-0. Microbiology (Reading). 2006. PMID: 16514151
-
The core metal-recognition domain of MerR.Biochemistry. 1998 Nov 10;37(45):15885-95. doi: 10.1021/bi9817562. Biochemistry. 1998. PMID: 9843394
-
Bacterial resistances to inorganic mercury salts and organomercurials.Plasmid. 1992 Jan;27(1):4-16. doi: 10.1016/0147-619x(92)90002-r. Plasmid. 1992. PMID: 1311113 Review.
-
The MerR family of transcriptional regulators.FEMS Microbiol Rev. 2003 Jun;27(2-3):145-63. doi: 10.1016/S0168-6445(03)00051-2. FEMS Microbiol Rev. 2003. PMID: 12829265 Review.
Cited by
-
Adsorption of Hg2+/Cr6+ by metal-binding proteins heterologously expressed in Escherichia coli.BMC Biotechnol. 2024 Mar 23;24(1):15. doi: 10.1186/s12896-024-00842-9. BMC Biotechnol. 2024. PMID: 38521922 Free PMC article.
-
Versatile artificial mer operons in Escherichia coli towards whole cell biosensing and adsorption of mercury.PLoS One. 2021 May 26;16(5):e0252190. doi: 10.1371/journal.pone.0252190. eCollection 2021. PLoS One. 2021. PMID: 34038487 Free PMC article.
-
Surface display of PbrR on Escherichia coli and evaluation of the bioavailability of lead associated with engineered cells in mice.Sci Rep. 2018 Apr 9;8(1):5685. doi: 10.1038/s41598-018-24134-3. Sci Rep. 2018. PMID: 29632327 Free PMC article.
References
-
- Ankudinov, A. L., C. E. Bouldin, J. J. Rehr, J. Sims, and H. Hung. 2002. Parallel calculation of electron multiple scattering using Lanczos algorithms. Phys. Rev. B 65:104107.1-104107.11.
-
- Barkay, T., S. M. Miller, and A. O. Summers. 2003. Bacterial mercury resistance from atoms to ecosystems. FEMS Microbiol. Rev. 27:355-384. - PubMed
-
- Bontidean, I., J. R. Lloyd, J. L. Hobman, J. R. Wilson, E. Csoregi, B. Mattiasson, and N. L. Brown. 2000. Bacterial metal-resistance proteins and their use in biosensors for the detection of bioavailable heavy metals. J. Inorg. Biochem. 79:225-229. - PubMed
-
- Brocklehurst, K. R., J. L. Hobman, B. Lawley, L. Blank, S. J. Marshall, N. L. Brown, and A. P. Morby. 1999. ZntR is a Zn(II)-responsive MerR-like transcriptional regulator of zntA in Escherichia coli. Mol. Microbiol. 31:893-902. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

