DNA interaction and phosphotransfer of the C4-dicarboxylate-responsive DcuS-DcuR two-component regulatory system from Escherichia coli
- PMID: 14996819
- PMCID: PMC355966
- DOI: 10.1128/JB.186.6.1879-1889.2004
DNA interaction and phosphotransfer of the C4-dicarboxylate-responsive DcuS-DcuR two-component regulatory system from Escherichia coli
Abstract
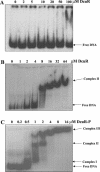
The DcuS-DcuR system of Escherichia coli is a two-component sensor-regulator that controls gene expression in response to external C(4)-dicarboxylates and citrate. The DcuS protein is particularly interesting since it contains two PAS domains, namely a periplasmic C(4)-dicarboxylate-sensing PAS domain (PASp) and a cytosolic PAS domain (PASc) of uncertain function. For a study of the role of the PASc domain, three different fragments of DcuS were overproduced and examined: they were PASc-kinase, PASc, and kinase. The two kinase-domain-containing fragments were autophosphorylated by [gamma-(32)P]ATP. The rate was not affected by fumarate or succinate, supporting the role of the PASp domain in C(4)-dicarboxylate sensing. Both of the phosphorylated DcuS constructs were able to rapidly pass their phosphoryl groups to DcuR, and after phosphorylation, DcuR dephosphorylated rapidly. No prosthetic group or significant quantity of metal was found associated with either of the PASc-containing proteins. The DNA-binding specificity of DcuR was studied by use of the pure protein. It was found to be converted from a monomer to a dimer upon acetylphosphate treatment, and native polyacrylamide gel electrophoresis suggested that it can oligomerize. DcuR specifically bound to the promoters of the three known DcuSR-regulated genes (dctA, dcuB, and frdA), with apparent K(D)s of 6 to 32 micro M for untreated DcuR and < or =1 to 2 microM for the acetylphosphate-treated form. The binding sites were located by DNase I footprinting, allowing a putative DcuR-binding motif [tandemly repeated (T/A)(A/T)(T/C)(A/T)AA sequences] to be identified. The DcuR-binding sites of the dcuB, dctA, and frdA genes were located 27, 94, and 86 bp, respectively, upstream of the corresponding +1 sites, and a new promoter was identified for dcuB that responds to DcuR.
Figures

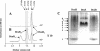




References
-
- Aiba, H., S. Adkya, and B. de Crombrugghe. 1981. Evidence for a functional gal promoter in intact Escherichia coli cells. J. Biol. Chem. 256:11905-11910. - PubMed
-
- Bartolomé, B., Y. Jubete, E. Martinez, and F. Delacruz. 1991. Construction and properties of a family of pACYC184-derived cloning vectors compatible with pBR322 and its derivatives. Gene 102:75-78. - PubMed
-
- Bott, M., M. Meyer, and P. Dimroth. 1995. Regulation of anaerobic citrate metabolism in Klebsiella pneumoniae. Mol. Microbiol. 18:533-546. - PubMed
-
- Bourret, R. B., K. A. Borkovich, and M. I. Simon. 1991. Signal transduction pathways involving protein phosphorylation in prokaryotes. Annu. Rev. Biochem. 60:401-441. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

