The eutD gene of Salmonella enterica encodes a protein with phosphotransacetylase enzyme activity
- PMID: 14996820
- PMCID: PMC355984
- DOI: 10.1128/JB.186.6.1890-1892.2004
The eutD gene of Salmonella enterica encodes a protein with phosphotransacetylase enzyme activity
Abstract
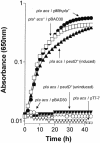
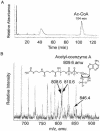
The EutD protein of Salmonella enterica is homologous to the catalytic domain of the phosphotransacetylase (Pta) enzyme. The Pta-like activity level of the EutD enzyme compared favorably to that of other Pta enzymes. High-pressure liquid chromatography and mass spectrometry verified that acetyl-coenzyme A was the product of the reaction. The EutD protein restored growth of an S. enterica pta strain on acetate as the source of carbon and energy.
Figures

References
-
- Babior, B. M. 1982. Ethanolamine ammonia-lyase, p. 263-288. In D. Dolphin (ed.), B12, vol. 2. John Wiley & Sons, New York, N.Y.
-
- Bandarian, V., R. R. Poyner, and G. H. Reed. 1999. Hydrogen atom exchange between 5′-deoxyadenosine and hydroxyethylhydrazine during the single turnover inactivation of ethanolamine ammonia-lyase. Biochemistry 38:12403-12407. - PubMed
-
- Bandarian, V., and G. H. Reed. 2002. Analysis of the electron paramagnetic resonance spectrum of a radical intermediate in the coenzyme B(12)-dependent ethanolamine ammonia-lyase catalyzed reaction of S-2-aminopropanol. Biochemistry 41:8580-8588. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

