Natural selection and molecular evolution in PTC, a bitter-taste receptor gene
- PMID: 14997422
- PMCID: PMC1181941
- DOI: 10.1086/383092
Natural selection and molecular evolution in PTC, a bitter-taste receptor gene
Abstract
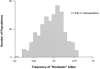
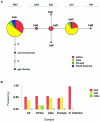
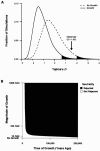
The ability to taste phenylthiocarbamide (PTC) is a classic phenotype that has long been known to vary in human populations. This phenotype is of genetic, epidemiologic, and evolutionary interest because the ability to taste PTC is correlated with the ability to taste other bitter substances, many of which are toxic. Thus, variation in PTC perception may reflect variation in dietary preferences throughout human history and could correlate with susceptibility to diet-related diseases in modern populations. To test R. A. Fisher's long-standing hypothesis that variability in PTC perception has been maintained by balancing natural selection, we examined patterns of DNA sequence variation in the recently identified PTC gene, which accounts for up to 85% of phenotypic variance in the trait. We analyzed the entire coding region of PTC (1,002 bp) in a sample of 330 chromosomes collected from African (n=62), Asian (n=138), European (n=110), and North American (n=20) populations by use of new statistical tests for natural selection that take into account the potentially confounding effects of human population growth. Two intermediate-frequency haplotypes corresponding to "taster" and "nontaster" phenotypes were found. These haplotypes had similar frequencies across Africa, Asia, and Europe. Genetic differentiation between the continental population samples was low (FST=0.056) in comparison with estimates based on other genes. In addition, Tajima's D and Fu and Li's D and F statistics demonstrated a significant deviation from neutrality because of an excess of intermediate-frequency variants when human population growth was taken into account (P<.01). These results combine to suggest that balancing natural selection has acted to maintain "taster" and "nontaster" alleles at the PTC locus in humans.
Figures

References
Electronic-Database Information
-
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for PTC) - PubMed
References
-
- Anonymous (1931) Tasteblindness. Science Suppl 73:14
-
- Barnicot NA, Harris H, Kalmus H (1951) Taste thresholds of further eighteen compounds and their correlation with PTC thresholds. Ann Eugen 16:119–128 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

