Structural basis for the function of a minimembrane protein subunit of yeast oligosaccharyltransferase
- PMID: 15001703
- PMCID: PMC374328
- DOI: 10.1073/pnas.0400512101
Structural basis for the function of a minimembrane protein subunit of yeast oligosaccharyltransferase
Abstract
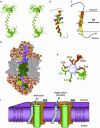
N-glycosylation of proteins is an essential, highly conserved modification reaction that occurs in all eukaryotes and some prokaryotes. This process is catalyzed by oligosaccharyltransferase (OT), a multisubunit enzyme localized in the endoplasmic reticulum. Complete loss of N-glycosylation is lethal in all organisms. In Saccharomyces cerevisiae, OT is composed of nine nonidentical membrane proteins. Here, we report the atomic structure of an OT subunit from S. cerevisiae, Ost4p. This unusually small membrane protein containing only 36 residues folds into a well formed, kinked helix in the model-membrane solvent system used in this study. The residues critical for the OT activity and the stability of Stt3p-Ost4p-Ost3p subcomplex are located in helix alpha2, the larger cytosolic half of this kinked helix. The residues known to disrupt Ost4p-Stt3p complex form a well defined ridge in the 3D structure. Taking together prior mutational studies and the NMR structure of Ost4p, we propose that in the OT complex Stt3p is packed against the alpha 2-helix of Ost4p by using a "ridges-into-grooves" model, with Met-18, Leu-21, and Ile-24 as the packing interface on one face, whereas Ost3p is involved in interactions with Met-19, Thr-20, Ile-22, and Val-23 on the other face.
Figures



References
-
- Wacker, M., Linton, D., Hitchen, P. G., Nita-Lazar, M., Haslam, S. M., North, S. J., Panico, M., Morris, H. R., Dell, A., Wren, B. W., et al. (2002) Science 298, 1790-1793. - PubMed
-
- Kornfeld, R. & Kornfeld, S. (1985) Annu. Rev. Biochem. 54, 631-664. - PubMed
-
- Knauer, R. & Lehle, L. (1999) Biochim. Biophys. Acta 1426, 256-273. - PubMed
-
- Marshall, R. D. (1972) Annu. Rev. Biochem. 41, 673-702. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

