Archaeal phylogeny based on proteins of the transcription and translation machineries: tackling the Methanopyrus kandleri paradox
- PMID: 15003120
- PMCID: PMC395767
- DOI: 10.1186/gb-2004-5-3-r17
Archaeal phylogeny based on proteins of the transcription and translation machineries: tackling the Methanopyrus kandleri paradox
Abstract
Background: Phylogenetic analysis of the Archaea has been mainly established by 16S rRNA sequence comparison. With the accumulation of completely sequenced genomes, it is now possible to test alternative approaches by using large sequence datasets. We analyzed archaeal phylogeny using two concatenated datasets consisting of 14 proteins involved in transcription and 53 ribosomal proteins (3,275 and 6,377 positions, respectively).
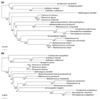
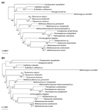
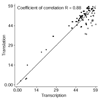
Results: Important relationships were confirmed, notably the dichotomy of the archaeal domain as represented by the Crenarchaeota and Euryarchaeota, the sister grouping of Sulfolobales and Aeropyrum pernix, and the monophyly of a large group comprising Thermoplasmatales, Archaeoglobus fulgidus, Methanosarcinales and Halobacteriales, with the latter two orders forming a robust cluster. The main difference concerned the position of Methanopyrus kandleri, which grouped with Methanococcales and Methanobacteriales in the translation tree, whereas it emerged at the base of the euryarchaeotes in the transcription tree. The incongruent placement of M. kandleri is likely to be the result of a reconstruction artifact due to the high evolutionary rates displayed by the components of its transcription apparatus.
Conclusions: We show that two informational systems, transcription and translation, provide a largely congruent signal for archaeal phylogeny. In particular, our analyses support the appearance of methanogenesis after the divergence of the Thermococcales and a late emergence of aerobic respiration from within methanogenic ancestors. We discuss the possible link between the evolutionary acceleration of the transcription machinery in M. kandleri and several unique features of this archaeon, in particular the absence of the elongation transcription factor TFS.
Figures
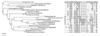


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

