FlyGEM, a full transcriptome array platform for the Drosophila community
- PMID: 15003122
- PMCID: PMC395769
- DOI: 10.1186/gb-2004-5-3-r19
FlyGEM, a full transcriptome array platform for the Drosophila community
Abstract
We have constructed a DNA microarray to monitor expression of predicted genes in Drosophila. By using homotypic hybridizations, we show that the array performs reproducibly, that dye effects are minimal, and that array results agree with systematic northern blotting. The array gene list has been extensively annotated and linked-out to other databases. Incyte and the NIH have made the platform available to the community via academic microarray facilities selected by an NIH committee.
Figures

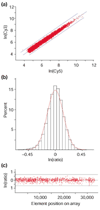

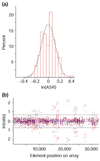
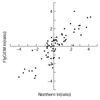
References
-
- Misra S, Crosby MA, Mungall CJ, Matthews BB, Campbell KS, Hradecky P, Huang Y, Kaminker JS, Millburn GH, Prochnik SE, et al. Annotation of the Drosophila melanogaster euchromatic genome: a systematic review. Genome Biol. 2002;3:research0083.1–0083.22. doi: 10.1186/gb-2002-3-12-research0083. - DOI - PMC - PubMed
-
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

