Preclinical evaluation of two real-time, reverse transcription-PCR assays for detection of the severe acute respiratory syndrome coronavirus
- PMID: 15004042
- PMCID: PMC356893
- DOI: 10.1128/JCM.42.3.987-991.2004
Preclinical evaluation of two real-time, reverse transcription-PCR assays for detection of the severe acute respiratory syndrome coronavirus
Abstract
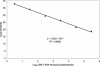
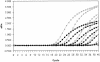
We verified the analytical performance characteristics of a previously described real-time reverse transcription-PCR (RT-PCR) assay targeting the open reading frame (ORF) 1b region of the severe acute respiratory syndrome coronavirus (SARS-CoV) with RNA transcripts. We then compared it to a novel nucleocapsid gene real-time RT-PCR assay with genomic RNA. The assays differed only in the primer and probe sequences and final concentrations. A commercially available armored RNA (Ambion, Austin, Tex.) was evaluated as positive control for the ORF 1b assay. The analytical sensitivity, reproducibility, amplification efficiency, and dynamic range of the assays were similar. Both were specific for SARS-CoV as determined by testing against human CoV 229E and OC43, specimens from patients without SARS, and by BLAST searches of GenBank for primer and probe sequence homology. The armored RNA was found to be a suitable positive control for the ORF 1b assay that could be reliably recovered and amplified from a variety of clinical specimens.
Figures

References
-
- Drosten, C., S. Gottig, S. Schilling, M. Asper, M. Panning, H. Schmitz, and S. Gunther. 2002. Rapid detection and quantification of RNA or Ebola and Marburg viruses, Lassa virus, Crimean-Congo hemorrhagic fever virus, Rift Valley fever virus, dengue virus, and yellow fever virus by real-time reverse transcription-PCR. J. Clin. Microbiol. 40:2323-2330. - PMC - PubMed
-
- Drosten, C., S. Gunther, W. Preiser, S. van der Werf, H. R. Brodt, S. Becker, H. Rabenau, M. Panning, L. Kolesnikova, R. A. Fouchier, A. Berger, A. M. Burguiere, J. Cinatl, M. Eickmann, N. Escriou, K. Grywna, S. Kramme, J. C. Manuguerra, S. Muller, V. Rickerts, M. Sturmer, S. Vieth, H. D. Klenk, A. D. Osterhaus, H. Schmitz, and H. W. Doerr. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 348:1967-1976. - PubMed
-
- Ksiazek, T. G., D. Erdman, C. S. Goldsmith, S. R. Zaki, T. Peret, S. Emery, S. Tong, C. Urbani, J. A. Comer, W. Lim, P. E. Rollin, S. F. Dowell, A. E. Ling, C. D. Humphrey, W. J. Shieh, J. Guarner, C. D. Paddock, P. Rota, B. Fields, J. DeRisi, J. Y. Yang, N. Cox, J. M. Hughes, J. W. LeDuc, W. J. Bellini, L. J. Anderson, and S. W. Group. 2003. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 348:1953-1966. - PubMed
-
- Marra, M. A., S. J. Jones, C. R. Astell, R. A. Holt, A. Brooks-Wilson, Y. S. Butterfield, J. Khattra, J. K. Asano, S. A. Barber, S. Y. Chan, A. Cloutier, S. M. Coughlin, D. Freeman, N. Girn, O. L. Griffith, S. R. Leach, M. Mayo, H. McDonald, S. B. Montgomery, P. K. Pandoh, A. S. Petrescu, A. G. Robertson, J. E. Schein, A. Siddiqui, D. E. Smailus, J. M. Stott, G. S. Yang, F. Plummer, A. Andonov, H. Artsob, N. Bastien, K. Bernard, T. F. Booth, D. Bowness, M. Czub, M. Drebot, L. Fernando, R. Flick, M. Garbutt, M. Gray, A. Grolla, S. Jones, H. Feldmann, A. Meyers, A. Kabani, Y. Li, S. Normand, U. Stroher, G. A. Tipples, S. Tyler, R. Vogrig, D. Ward, B. Watson, R. C. Brunham, M. Krajden, M. Petric, D. M. Skowronski, C. Upton, and R. L. Roper. 2003. The genome sequence of the SARS-associated coronavirus. Science 300:1399-1404. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

