Optimization of quantitative detection of cytomegalovirus DNA in plasma by real-time PCR
- PMID: 15004066
- PMCID: PMC356812
- DOI: 10.1128/JCM.42.3.1142-1148.2004
Optimization of quantitative detection of cytomegalovirus DNA in plasma by real-time PCR
Abstract
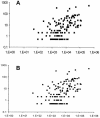
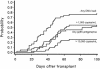
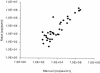
Previous studies have shown that detection of cytomegalovirus (CMV) DNA in plasma is less sensitive than the antigenemia assay for CMV surveillance in blood. In 1,983 blood samples, plasma PCR assays with three different primer sets (UL125 alone, UL126 alone, and UL55/UL123-exon 4) were compared to the pp65 antigenemia assay and blood cultures. Plasma PCR detected CMV more frequently in blood specimens than either the antigenemia assay or cultures, but of the three PCR assays, the double-primer assay (UL55/UL123-exon 4) performed best with regard to sensitivity, specificity, and predictive values compared to antigenemia: 122 of 151 antigenemia-positive samples were detected (sensitivity, 80.1%), and there were 122 samples that were PCR positive-antigenemia negative (specificity, 93%). Samples with discrepant results had a low viral load (median, 0.5 cells per slide; 1,150 copies per ml) and were often obtained from patients receiving antiviral therapy. CMV could be detected by other methods in 15 of 29 antigenemia positive-PCR negative samples compared to 121 of 122 PCR positive-antigenemia negative samples (P < 0.001). On a per-subject basis, 21 of 25 patients (antigenemia positive-PCR negative) and all 57 (PCR positive-antigenemia negative) could be confirmed at different time points during follow-up. The higher sensitivity of the double-primer assay resulted in earlier detection compared to antigenemia in a time-to-event analysis of 42 CMV-seropositive stem cell transplant recipients, and two of three patients with CMV disease who were antigenemia negative were detected by plasma PCR prior to the onset of disease. Interassay variability was low, and the dynamic range was >5 log(10). Automated DNA extraction resulted in high reproducibility, accurate CMV quantitation (R = 0.87, P < 0.001), improved sensitivity, and increased speed of sample processing. Thus, primer optimization and improved DNA extraction techniques resulted in a plasma-based PCR assay that is significantly more sensitive than pp65 antigenemia and blood cultures for detection of CMV in blood specimens.
Figures

References
-
- Berger, A., J. Braner, H. W. Doerr, and B. Weber. 1998. Quantification of viral load: clinical relevance for human immunodeficiency virus, hepatitis B virus, and hepatitis C virus infection. Intervirology 41:24-34. - PubMed
-
- Boeckh, M., G. M. Gallez-Hawkins, D. Myerson, J. A. Zaia, and R. A. Bowden. 1997. Plasma polymerase chain reaction for cytomegalovirus DNA after allogeneic marrow transplantation: comparison with polymerase chain reaction using peripheral blood leukocytes, pp65 antigenemia, and viral culture. Transplantation 64:108-113. - PubMed
-
- Boeckh, M., W. Leisenring, S. R. Riddell, R. A. Bowden, M. Huang, D. Myerson, T. Stevens-Ayers, M. E. D. Flowers, T. Cunningham, and L. Corey. 2003. Late cytomegalovirus disease and mortality in allogeneic hematopoietic stem cell transplant recipients: importance of viral load and T cell immunity. Blood 101:407-414. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

