DNA dynamically directs its own transcription initiation
- PMID: 15004245
- PMCID: PMC390311
- DOI: 10.1093/nar/gkh335
DNA dynamically directs its own transcription initiation
Abstract
It has long been known that double-stranded DNA is subject to temporary, localized openings of its two strands. Particular regions along a DNA polymer are destabilized structurally by available thermal energy in the system. The localized sequence of DNA determines the physical properties of a stretch of DNA, and that in turn determines the opening profile of that DNA fragment. We show that the Peyrard-Bishop nonlinear dynamical model of DNA, which has been used to simulate denaturation of short DNA fragments, gives an accurate representation of the instability profile of a defined sequence of DNA, as verified using S1 nuclease cleavage assays. By comparing results for a non-promoter DNA fragment, the adenovirus major late promoter, the adeno-associated viral P5 promoter and a known P5 mutant promoter that is inactive for transcription, we show that the predicted openings correlate almost exactly with the promoter transcriptional start sites and major regulatory sites. Physicists have speculated that localized melting of DNA might play a role in gene transcription and other processes. Our data link sequence-dependent opening behavior in DNA to transcriptional activity for the first time.
Figures
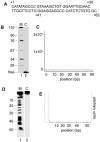
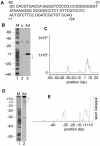

References
-
- Benham C.J. (1992) Energetics of the strand separation transition in superhelical DNA. J. Mol. Biol., 225, 835–847. - PubMed
-
- Benham C.J. (1996) Duplex destabilization in superhelical DNA is predicted to occur at specific transcriptional regulatory regions. J. Mol. Biol., 255, 425–434. - PubMed
-
- Packer M.J., Dauncey,M.P. and Hunter,C.A. (2000) Sequence-dependent DNA structure: tetranucleotide conformational maps. J. Mol. Biol., 295, 85–103. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

