Pattern of distribution and cycling of SLOB, Slowpoke channel binding protein, in Drosophila
- PMID: 15005796
- PMCID: PMC343274
- DOI: 10.1186/1471-2202-5-3
Pattern of distribution and cycling of SLOB, Slowpoke channel binding protein, in Drosophila
Abstract
Background: SLOB binds to and modulates the activity of the Drosophila Slowpoke (dSlo) calcium activated potassium channel. Recent microarray analyses demonstrated circadian cycling of slob mRNA.
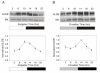
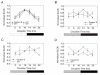
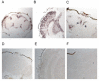
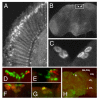
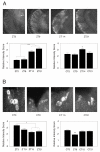
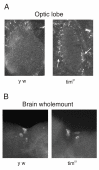
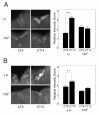
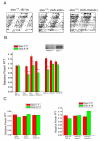
Results: We report the mRNA and protein expression pattern of slob in Drosophila heads. slob transcript is present in the photoreceptors, optic lobe, pars intercerebralis (PI) neurons and surrounding brain cortex. SLOB protein exhibits a similar distribution pattern, and we show that it cycles in Drosophila heads, in photoreceptor cells and in neurosecretory cells of the PI. The cycling of SLOB is altered in various clock gene mutants, and SLOB is expressed in ectopic locations in tim01 flies. We also demonstrate that SLOB no longer cycles in the PI neurons of Clkjrk flies, and that SLOB expression is reduced in the PI neurons of flies that lack pigment dispersing factor (PDF), a neuropeptide secreted by clock cells.
Conclusions: These data are consistent with the idea that SLOB may participate in one or more circadian pathways in Drosophila.
Figures
References
-
- Claridge-Chang A, Wijnen H, Naef F, Boothroyd C, Rajewsky N, Young MW. Circadian regulation of gene expression systems in the Drosophila head. Neuron. 2001;32:657–671. - PubMed
-
- McDonald MJ, Rosbash M. Microarray analysis and organization of circadian gene expression in Drosophila. Cell. 2001;107:567–578. - PubMed
-
- Lin Y, Han M, Shimada B, Wang L, Gibler TM, Amarakone A, Awad TA, Stormo GD, Van Gelder RN, Taghert PH. Influence of the period-dependent circadian clock on diurnal, circadian, and aperiodic gene expression in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2002;99:9562–9567. doi: 10.1073/pnas.132269699. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

