Aerobic and anaerobic toluene degradation by a newly isolated denitrifying bacterium, Thauera sp. strain DNT-1
- PMID: 15006757
- PMCID: PMC368410
- DOI: 10.1128/AEM.70.3.1385-1392.2004
Aerobic and anaerobic toluene degradation by a newly isolated denitrifying bacterium, Thauera sp. strain DNT-1
Abstract
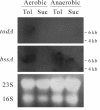
A newly isolated denitrifying bacterium, Thauera sp. strain DNT-1, grew on toluene as the sole carbon and energy source under both aerobic and anaerobic conditions. When this strain was cultivated under oxygen-limiting conditions with nitrate, first toluene was degraded as oxygen was consumed, while later toluene was degraded as nitrate was reduced. Biochemical observations indicated that initial degradation of toluene occurred through a dioxygenase-mediated pathway and the benzylsuccinate pathway under aerobic and denitrifying conditions, respectively. Homologous genes for toluene dioxygenase (tod) and benzylsuccinate synthase (bss), which are the key enzymes in aerobic and anaerobic toluene degradation, respectively, were cloned from genomic DNA of strain DNT-1. The results of Northern blot analyses and real-time quantitative reverse transcriptase PCR suggested that transcription of both sets of genes was induced by toluene. In addition, the tod genes were induced under aerobic conditions, whereas the bss genes were induced under both aerobic and anaerobic conditions. On the basis of these results, it is concluded that strain DNT-1 modulates the expression of two different initial pathways of toluene degradation according to the availability of oxygen in the environment.
Figures


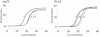
References
-
- Assinder, S. J., and P. A. Williams. 1990. The TOL plasmids: determinants of the catabolism of toluene and xylenes. Adv. Microb. Physiol. 31:1-69. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

