Telomere-bound TRF1 and TRF2 stall the replication fork at telomeric repeats
- PMID: 15007108
- PMCID: PMC390322
- DOI: 10.1093/nar/gkh309
Telomere-bound TRF1 and TRF2 stall the replication fork at telomeric repeats
Abstract
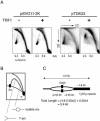
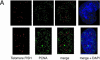
Vertebrate telomeres consist of tandem repeats of T2AG3 and associated proteins including the telomeric DNA-binding proteins, TRF1 and TRF2. It has been proposed that telomeres assume two interswitchable states, the open state that is accessible to various trans-acting factors and the closed state that excludes those factors. TRF1 and TRF2 are believed to promote the formation of the closed state. However, little is known about how those two states influence DNA replication. We analyzed the effects of TRF1 and TRF2 on telomeric replication both in vitro and in vivo. By exploiting the in vitro replication system of linear SV40 DNA, we found that telomeric repeats are a poor replication template. Moreover, the addition of recombinant TRF1 and TRF2 significantly stalled the replication fork progression at telomeric repeats. When TRF1 was overexpressed in HeLa cells, cells with 4N DNA content were accumulated. Furthermore, cytological analyses revealed that the replication focus overlapped with telomere signals at a significantly higher frequency in TRF1-overexpressing cells than in control cells. The results suggest that TRF1 and TRF2 exert inhibitory effects on replication fork progression.
Figures





Similar articles
-
[Recognition of internal (TTAGGG)n repeats by telomeric protein TRF1 and its role in maintenance of chromosomal stability in Chinese hamster cells].Tsitologiia. 2003;45(12):1211-20. Tsitologiia. 2003. PMID: 15027354 Russian.
-
Oxidative damage in telomeric DNA disrupts recognition by TRF1 and TRF2.Nucleic Acids Res. 2005 Feb 24;33(4):1230-9. doi: 10.1093/nar/gki273. Print 2005. Nucleic Acids Res. 2005. PMID: 15731343 Free PMC article.
-
Human telomeres contain two distinct Myb-related proteins, TRF1 and TRF2.Nat Genet. 1997 Oct;17(2):231-5. doi: 10.1038/ng1097-231. Nat Genet. 1997. PMID: 9326950
-
[Regulation of telomeres length: making the telomeres accessible?].Bull Cancer. 2005 Jan;92(1):13-22. Bull Cancer. 2005. PMID: 15811846 Review. French.
-
Post-translational modifications of TRF1 and TRF2 and their roles in telomere maintenance.Mech Ageing Dev. 2012 Jun;133(6):421-34. doi: 10.1016/j.mad.2012.05.002. Epub 2012 May 23. Mech Ageing Dev. 2012. PMID: 22634377 Review.
Cited by
-
Disruption of Wnt/β-Catenin Signaling and Telomeric Shortening Are Inextricable Consequences of Tankyrase Inhibition in Human Cells.Mol Cell Biol. 2015 Jul;35(14):2425-35. doi: 10.1128/MCB.00392-15. Epub 2015 May 4. Mol Cell Biol. 2015. PMID: 25939383 Free PMC article.
-
Solving the Telomere Replication Problem.Genes (Basel). 2017 Jan 31;8(2):55. doi: 10.3390/genes8020055. Genes (Basel). 2017. PMID: 28146113 Free PMC article. Review.
-
c-Myc interacts with TRF1/PIN2 and regulates telomere length.Biochem Biophys Res Commun. 2007 Nov 3;362(4):842-7. doi: 10.1016/j.bbrc.2007.08.064. Epub 2007 Aug 22. Biochem Biophys Res Commun. 2007. PMID: 17765874 Free PMC article.
-
Fusion of short telomeres in human cells is characterized by extensive deletion and microhomology, and can result in complex rearrangements.Nucleic Acids Res. 2010 Apr;38(6):1841-52. doi: 10.1093/nar/gkp1183. Epub 2009 Dec 21. Nucleic Acids Res. 2010. PMID: 20026586 Free PMC article.
-
TRF1 relies on fork reversal to prevent fragility at human telomeres.Nat Commun. 2025 Jul 11;16(1):6439. doi: 10.1038/s41467-025-61828-5. Nat Commun. 2025. PMID: 40645989 Free PMC article.
References
-
- Chong L., van Steensel,B., Broccoli,D., Erdjument,B.H., Hanish,J., Tempst,P. and de Lange,T. (1995) A human telomeric protein. Science, 270, 1663–1667. - PubMed
-
- Broccoli D., Smogorzewska,A., Chong,L. and de Lange,T. (1997) Human telomeres contain two distinct Myb-related proteins, TRF1 and TRF2. Nature Genet., 17, 231–235. - PubMed
-
- van Steensel B. and de Lange,T. (1997) Control of telomere length by the human telomeric protein TRF1. Nature, 385, 740–743. - PubMed
-
- Griffith J.D., Comeau,L., Rosenfield,S., Stansel,R.M., Bianchi,A., Moss,H. and de Lange,T. (1999) Mammalian telomeres end in a large duplex loop. Cell, 97, 503–514. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

