Microsecond folding dynamics of the F13W G29A mutant of the B domain of staphylococcal protein A by laser-induced temperature jump
- PMID: 15007169
- PMCID: PMC374326
- DOI: 10.1073/pnas.0306433101
Microsecond folding dynamics of the F13W G29A mutant of the B domain of staphylococcal protein A by laser-induced temperature jump
Abstract
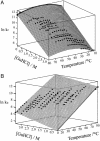
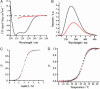
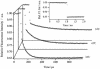
The small size (58 residues) and simple structure of the B domain of staphylococcal protein A (BdpA) have led to this domain being a paradigm for theoretical studies of folding. Experimental studies of the folding of BdpA have been limited by the rapidity of its folding kinetics. We report the folding kinetics of a fluorescent mutant of BdpA (G29A F13W), named F13W*, using nanosecond laser-induced temperature jump experiments. Automation of the apparatus has permitted large data sets to be acquired that provide excellent signal-to-noise ratio over a wide range of experimental conditions. By measuring the temperature and denaturant dependence of equilibrium and kinetic data for F13W*, we show that thermodynamic modeling of multidimensional equilibrium and kinetic surfaces is a robust method that allows reliable extrapolation of rate constants to regions of the folding landscape not directly accessible experimentally. The results reveal that F13W* is the fastest-folding protein of its size studied to date, with a maximum folding rate constant at 0 M guanidinium chloride and 45 degrees C of 249,000 s(-1). Assuming the single-exponential kinetics represent barrier-limited folding, these data limit the value for the preexponential factor for folding of this protein to at least approximately 2 x 10(6) s(-1).
Figures


References
-
- Jackson, S. E. & Fersht, A. R. (1991) Biochemistry 30, 10428-10435. - PubMed
-
- Kragelund, B. B., Robinson, C. V., Knudsen, J., Dobson, C. M. & Poulsen, F. M. (1995) Biochemistry 34, 7217-7224. - PubMed
-
- Villegas, V., Azuaga, A., Catasus, L., Reverter, D., Mateo, P. L., Aviles, F. X. & Serrano, L. (1995) Biochemistry 34, 15105-15110. - PubMed
-
- Schindler, T., Herrler, M., Marahiel, M. A. & Schmid, F. X. (1995) Nat. Struct. Biol. 2, 663-673. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

