Structure of a thrombospondin C-terminal fragment reveals a novel calcium core in the type 3 repeats
- PMID: 15014436
- PMCID: PMC381422
- DOI: 10.1038/sj.emboj.7600166
Structure of a thrombospondin C-terminal fragment reveals a novel calcium core in the type 3 repeats
Abstract
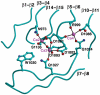
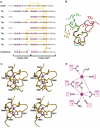
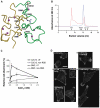
Thrombospondins (TSPs) are extracellular regulators of cell-matrix interactions and cell phenotype. The most highly conserved region of all TSPs are the calcium-binding type 3 (T3) repeats and the C-terminal globular domain (CTD). The crystal structure of a cell-binding TSP-1 fragment, spanning three T3 repeats and the CTD, reveals a compact assembly. The T3 repeats lack secondary structure and are organised around a core of calcium ions; two DxDxDGxxDxxD motifs per repeat each encapsulate two calcium ions in a novel arrangement. The CTD forms a lectin-like beta-sandwich and contains four strictly conserved calcium-binding sites. Disruption of the hairpin structure of T3 repeats 6 and 7 decreases protein secretion and stability. The availability for cell attachment of an RGD motif in T3 repeat 7 is modulated by calcium loading. The central architectural role of calcium explains how it is critical for the functions of the TSP C-terminal region. Mutations in the T3 repeats of TSP-5/COMP, which cause two human skeletal disorders, are predicted to disrupt the tertiary structure of the T3-CTD assembly.
Figures



References
-
- Adams JC (2001) Thrombospondins: multifunctional regulators of cell interactions. Annu Rev Cell Dev Biol 17: 25–51 - PubMed
-
- Adams JC, Lawler J (1993a) The thrombospondin family. Curr Biol 3: 188–190 - PubMed
-
- Adams JC, Lawler J (1993b) Diverse mechanisms for cell attachment to platelet thrombospondin. J Cell Sci 104: 1061–1071 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

