Hierarchical targeting of subtype C human immunodeficiency virus type 1 proteins by CD8+ T cells: correlation with viral load
- PMID: 15016844
- PMCID: PMC371059
- DOI: 10.1128/jvi.78.7.3233-3243.2004
Hierarchical targeting of subtype C human immunodeficiency virus type 1 proteins by CD8+ T cells: correlation with viral load
Abstract
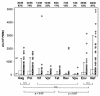
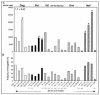
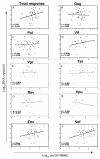
An understanding of the relationship between the breadth and magnitude of T-cell epitope responses and viral loads is important for the design of effective vaccines. For this study, we screened a cohort of 46 subtype C human immunodeficiency virus type 1 (HIV-1)-infected individuals for T-cell responses against a panel of peptides corresponding to the complete subtype C genome. We used a gamma interferon ELISPOT assay to explore the hypothesis that patterns of T-cell responses across the expressed HIV-1 genome correlate with viral control. The estimated median time from seroconversion to response for the cohort was 13 months, and the order of cumulative T-cell responses against HIV proteins was as follows: Nef > Gag > Pol > Env > Vif > Rev > Vpr > Tat > Vpu. Nef was the most intensely targeted protein, with 97.5% of the epitopes being clustered within 119 amino acids, constituting almost one-third of the responses across the expressed genome. The second most targeted region was p24, comprising 17% of the responses. There was no correlation between viral load and the breadth of responses, but there was a weak positive correlation (r = 0.297; P = 0.034) between viral load and the total magnitude of responses, implying that the magnitude of T-cell recognition did not contribute to viral control. When hierarchical patterns of recognition were correlated with the viral load, preferential targeting of Gag was significantly (r = 0.445; P = 0.0025) associated with viral control. These data suggest that preferential targeting of Gag epitopes, rather than the breadth or magnitude of the response across the genome, may be an important marker of immune efficacy. These data have significance for the design of vaccines and for interpretation of vaccine-induced responses.
Figures



Similar articles
-
Human immunodeficiency virus-specific gamma interferon enzyme-linked immunospot assay responses targeting specific regions of the proteome during primary subtype C infection are poor predictors of the course of viremia and set point.J Virol. 2009 Jan;83(1):470-8. doi: 10.1128/JVI.01678-08. Epub 2008 Oct 22. J Virol. 2009. PMID: 18945774 Free PMC article.
-
Role of human immunodeficiency virus (HIV)-specific T-cell immunity in control of dual HIV-1 and HIV-2 infection.J Virol. 2007 Sep;81(17):9061-71. doi: 10.1128/JVI.00117-07. Epub 2007 Jun 20. J Virol. 2007. PMID: 17582003 Free PMC article.
-
HIV-1 superinfection despite broad CD8+ T-cell responses containing replication of the primary virus.Nature. 2002 Nov 28;420(6914):434-9. doi: 10.1038/nature01200. Nature. 2002. PMID: 12459786
-
CD8+ cells in human immunodeficiency virus type I pathogenesis: cytolytic and noncytolytic inhibition of viral replication.Adv Immunol. 1997;66:273-311. doi: 10.1016/s0065-2776(08)60600-8. Adv Immunol. 1997. PMID: 9328644 Review. No abstract available.
-
Regulatory and accessory HIV-1 proteins: potential targets for HIV-1 vaccines?Curr Med Chem. 2005;12(6):741-7. doi: 10.2174/0929867053202205. Curr Med Chem. 2005. PMID: 15790309 Review.
Cited by
-
Early Gag immunodominance of the HIV-specific T-cell response during acute/early infection is associated with higher CD8+ T-cell antiviral activity and correlates with preservation of the CD4+ T-cell compartment.J Virol. 2013 Jul;87(13):7445-62. doi: 10.1128/JVI.00865-13. Epub 2013 Apr 24. J Virol. 2013. PMID: 23616666 Free PMC article.
-
Clinical Control of HIV-1 by Cytotoxic T Cells Specific for Multiple Conserved Epitopes.J Virol. 2015 May;89(10):5330-9. doi: 10.1128/JVI.00020-15. Epub 2015 Mar 4. J Virol. 2015. PMID: 25741000 Free PMC article.
-
Influence of HAART on alternative reading frame immune responses over the course of HIV-1 infection.PLoS One. 2012;7(6):e39311. doi: 10.1371/journal.pone.0039311. Epub 2012 Jun 29. PLoS One. 2012. PMID: 22768072 Free PMC article.
-
Early HLA-B*57-restricted CD8+ T lymphocyte responses predict HIV-1 disease progression.J Virol. 2012 Oct;86(19):10505-16. doi: 10.1128/JVI.00102-12. Epub 2012 Jul 18. J Virol. 2012. PMID: 22811521 Free PMC article.
-
Early induction of polyfunctional simian immunodeficiency virus (SIV)-specific T lymphocytes and rapid disappearance of SIV from lymph nodes of sooty mangabeys during primary infection.J Immunol. 2011 May 1;186(9):5151-61. doi: 10.4049/jimmunol.1004110. Epub 2011 Mar 25. J Immunol. 2011. PMID: 21441446 Free PMC article.
References
-
- Addo, M. M., X. G. Yu, A. Rathod, D. Cohen, R. L. Eldridge, D. Strick, M. N. Johnston, C. Corcoran, A. G. Wurcel, C. A. Fitzpatrick, M. E. Feeney, W. R. Rodriguez, N. Basgoz, R. Draenert, D. R. Stone, C. Brander, P. J. Goulder, E. S. Rosenberg, M. Altfeld, and B. D. Walker. 2003. Comprehensive epitope analysis of human immunodeficiency virus type 1 (HIV-1)-specific T-cell responses directed against the entire expressed HIV-1 genome demonstrate broadly directed responses, but no correlation to viral load. J. Virol. 77:2081-2092. - PMC - PubMed
-
- Altfeld, M., M. M. Addo, R. Shankarappa, P. K. Lee, T. M. Allen, X. G. Yu, A. Rathod, J. Harlow, K. O'Sullivan, M. N. Johnston, P. J. Goulder, J. I. Mullins, E. S. Rosenberg, C. Brander, B. Korber, and B. D. Walker. 2003. Enhanced detection of human immunodeficiency virus type 1-specific T-cell responses to highly variable regions by using peptides based on autologous virus sequences. J. Virol. 77:7330-7340. - PMC - PubMed
-
- Amara, R. R., F. Villinger, J. D. Altman, S. L. Lydy, S. P. O'Neil, S. I. Staprans, D. C. Montefiori, Y. Xu, J. G. Herndon, L. S. Wyatt, M. A. Candido, N. L. Kozyr, P. L. Earl, J. M. Smith, H. L. Ma, B. D. Grimm, M. L. Hulsey, J. Miller, H. M. McClure, J. M. McNicholl, B. Moss, and H. L. Robinson. 2001. Control of a mucosal challenge and prevention of AIDS by a multiprotein DNA/MVA vaccine. Science 292:69-74. - PubMed
-
- Barouch, D. H., S. Santra, M. J. Kuroda, J. E. Schmitz, R. Plishka, A. Buckler-White, A. E. Gaitan, R. Zin, J. H. Nam, L. S. Wyatt, M. A. Lifton, C. E. Nickerson, B. Moss, D. C. Montefiori, V. M. Hirsch, and N. L. Letvin. 2001. Reduction of simian-human immunodeficiency virus 89.6P viremia in rhesus monkeys by recombinant modified vaccinia virus Ankara vaccination. J. Virol. 75:5151-5158. - PMC - PubMed
-
- Barouch, D. H., S. Santra, J. E. Schmitz, M. J. Kuroda, T. M. Fu, W. Wagner, M. Bilska, A. Craiu, X. X. Zheng, G. R. Krivulka, K. Beaudry, M. A. Lifton, C. E. Nickerson, W. L. Trigona, K. Punt, D. C. Freed, L. Guan, S. Dubey, D. Casimiro, A. Simon, M. E. Davies, M. Chastain, T. B. Strom, R. S. Gelman, D. C. Montefiori, M. G. Lewis, E. A. Emini, J. W. Shiver, and N. L. Letvin. 2000. Control of viremia and prevention of clinical AIDS in rhesus monkeys by cytokine-augmented DNA vaccination. Science 290:486-492. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

