Alternative approaches for efficient inhibition of hepatitis C virus RNA replication by small interfering RNAs
- PMID: 15016866
- PMCID: PMC371081
- DOI: 10.1128/jvi.78.7.3436-3446.2004
Alternative approaches for efficient inhibition of hepatitis C virus RNA replication by small interfering RNAs
Abstract
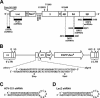
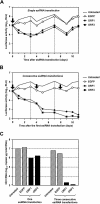
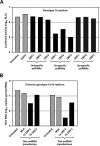
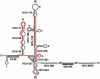
Persistent infection with hepatitis C virus (HCV) is a leading cause of chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma. It has recently been shown that HCV RNA replication is susceptible to small interfering RNAs (siRNAs), but the antiviral activity of siRNAs depends very much on their complementarity to the target sequence. Thus, the high degree of sequence diversity between different HCV genotypes and the rapid evolution of new quasispecies is a major problem in the development of siRNA-based gene therapies. For this study, we developed two alternative strategies to overcome these obstacles. In one approach, we used endoribonuclease-prepared siRNAs (esiRNAs) to simultaneously target multiple sites of the viral genome. We show that esiRNAs directed against various regions of the HCV coding sequence as well as the 5' nontranslated region (5' NTR) efficiently block the replication of subgenomic and genomic HCV replicons. In an alternative approach, we generated pseudotyped retroviruses encoding short hairpin RNAs (shRNAs). A total of 12 shRNAs, most of them targeting highly conserved sequence motifs within the 5' NTR or the early core coding region, were analyzed for their antiviral activities. After the transduction of Huh-7 cells containing a subgenomic HCV replicon, we found that all shRNAs targeting sequences in domain IV or nearby coding sequences blocked viral replication. In contrast, only one of seven shRNAs targeting sequences in domain II or III had a similar degree of antiviral activity, indicating that large sections of the NTRs are resistant to RNA interference. Moreover, we show that naive Huh-7 cells that stably expressed certain 5' NTR-specific shRNAs were largely resistant to a challenge with HCV replicons. These results demonstrate that the retroviral transduction of HCV-specific shRNAs provides a new possibility for antiviral intervention.
Figures



Similar articles
-
Hairpin ribozymes in combination with siRNAs against highly conserved hepatitis C virus sequence inhibit RNA replication and protein translation from hepatitis C virus subgenomic replicons.FEBS J. 2005 Nov;272(22):5910-22. doi: 10.1111/j.1742-4658.2005.04986.x. FEBS J. 2005. PMID: 16279954
-
Inhibition of hepatitis C virus infection in cell culture by small interfering RNAs.Mol Ther. 2007 Aug;15(8):1452-62. doi: 10.1038/sj.mt.6300186. Epub 2007 May 15. Mol Ther. 2007. PMID: 17505476 Free PMC article.
-
Suppression of hepatitis C virus replicon by RNA interference directed against the NS3 and NS5B regions of the viral genome.Microbiol Immunol. 2004;48(8):591-8. doi: 10.1111/j.1348-0421.2004.tb03556.x. Microbiol Immunol. 2004. PMID: 15322339
-
[Replication of hepatitis C virus genome].Uirusu. 2008 Dec;58(2):191-8. Uirusu. 2008. PMID: 19374197 Review. Japanese.
-
Potential role of RNAi in the treatment of HCV infection.Expert Rev Anti Infect Ther. 2007 Oct;5(5):823-31. doi: 10.1586/14787210.5.5.823. Expert Rev Anti Infect Ther. 2007. PMID: 17914916 Review.
Cited by
-
Criteria for effective design, construction, and gene knockdown by shRNA vectors.BMC Biotechnol. 2006 Jan 24;6:7. doi: 10.1186/1472-6750-6-7. BMC Biotechnol. 2006. PMID: 16433925 Free PMC article.
-
Endoribonuclease-prepared short interfering RNAs induce effective and specific inhibition of human immunodeficiency virus type 1 replication.J Virol. 2007 Oct;81(19):10680-6. doi: 10.1128/JVI.00950-07. Epub 2007 Jul 25. J Virol. 2007. PMID: 17652404 Free PMC article.
-
Inhibition of hepatitis C virus replication by intracellular delivery of multiple siRNAs by nanosomes.Mol Ther. 2012 Sep;20(9):1724-1736. doi: 10.1038/mt.2012.107. Epub 2012 May 22. Mol Ther. 2012. Retraction in: Mol Ther. 2025 Jan 8;33(1):422. doi: 10.1016/j.ymthe.2024.11.036. PMID: 22617108 Free PMC article. Retracted.
-
Sigma-1 receptor regulates early steps of viral RNA replication at the onset of hepatitis C virus infection.J Virol. 2013 Jun;87(11):6377-90. doi: 10.1128/JVI.03557-12. Epub 2013 Mar 27. J Virol. 2013. PMID: 23536676 Free PMC article.
-
Inhibition of HCV 5'-NTR and core expression by a small hairpin RNA delivered by a histone gene carrier, HPhA.Int J Med Sci. 2013 Jun 9;10(8):957-64. doi: 10.7150/ijms.5632. Print 2013. Int J Med Sci. 2013. PMID: 23801881 Free PMC article.
References
-
- Alt, M., R. Renz, P. H. Hofschneider, G. Paumgartner, and W. H. Caselmann. 1995. Specific inhibition of hepatitis C viral gene expression by antisense phosphorothioate oligodeoxynucleotides. Hepatology 22:707-717. - PubMed
-
- Ambros, V. 2003. MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell 113:673-676. - PubMed
-
- Bartenschlager, R., M. Frese, and T. Pietschmann. Novel insights into hepatitis C virus replication and persistence. Adv. Virus Res., in press. - PubMed
-
- Berberich-Siebelt, F., S. Klein-Hessling, N. Hepping, B. Santner-Nanan, D. Lindemann, A. Schimpl, I. Berberich, and E. Serfling. 2000. C/EBPbeta enhances IL-4 but impairs IL-2 and IFN-gamma induction in T cells. Eur. J. Immunol. 30:2576-2585. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

