High-resolution phylogenetic analysis of hepatitis C virus adaptation and its relationship to disease progression
- PMID: 15016867
- PMCID: PMC371055
- DOI: 10.1128/jvi.78.7.3447-3454.2004
High-resolution phylogenetic analysis of hepatitis C virus adaptation and its relationship to disease progression
Abstract
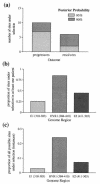
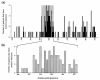
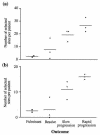
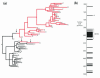
Hepatitis C virus (HCV) persists in the majority of those infected despite host immune responses. Evidence has accrued that selectively fixed mutations in the envelope genes (E1 and E2) are associated with viral persistence, particularly those that occur within the first hypervariable region of E2 (HVR1). However, the individual amino acid residues under selection have not been identified, nor have their selection pressures been measured, despite the importance of this information for understanding disease pathogenesis and for vaccine design. We performed a high-resolution analysis of published gene sequence data from individuals undergoing acute HCV infection, employing two phylogenetic methods to determine site-specific selection pressures. Strikingly, we found a statistically significant association between the number of sites selected and disease outcome, with the fewest selected sites in fulminant HCV cases and the greatest number of selected sites in rapid progressors, reflecting the duration and intensity of the arms race between host and virus. Moreover, sites outside the HVR1 appear to play a major role in viral evolution and pathogenesis, although there was no association between viral persistence and specific mutations in E1 and E2. Our analysis therefore allows fine dissection of immune selection pressures, which may be more diverse than previously thought. Such analyses could play a similarly informative role in studies of other persistent virus infections, such as human immunodeficiency virus.
Figures
Similar articles
-
Evolutionary study of hepatitis C virus envelope genes during primary infection.Chin Med J (Engl). 2007 Dec 20;120(24):2174-80. Chin Med J (Engl). 2007. PMID: 18167197
-
Cross-genotype characterization of genetic diversity and molecular adaptation in hepatitis C virus envelope glycoprotein genes.J Gen Virol. 2007 Feb;88(Pt 2):458-469. doi: 10.1099/vir.0.82357-0. J Gen Virol. 2007. PMID: 17251563
-
The outcome of acute hepatitis C predicted by the evolution of the viral quasispecies.Science. 2000 Apr 14;288(5464):339-44. doi: 10.1126/science.288.5464.339. Science. 2000. PMID: 10764648
-
Structure and Function of the Hepatitis C Virus Envelope Glycoproteins E1 and E2: Antiviral and Vaccine Targets.ACS Infect Dis. 2016 Nov 11;2(11):749-762. doi: 10.1021/acsinfecdis.6b00110. Epub 2016 Aug 16. ACS Infect Dis. 2016. PMID: 27933781 Review.
-
Genetic heterogeneity of hepatitis C virus: quasispecies and genotypes.Semin Liver Dis. 1995 Feb;15(1):41-63. doi: 10.1055/s-2007-1007262. Semin Liver Dis. 1995. PMID: 7597443 Review.
Cited by
-
Hepatitis C virus envelope glycoprotein co-evolutionary dynamics during chronic hepatitis C.Virology. 2008 Jun 5;375(2):580-91. doi: 10.1016/j.virol.2008.02.012. Epub 2008 Mar 17. Virology. 2008. PMID: 18343477 Free PMC article.
-
Dynamic features of the selective pressure on the human immunodeficiency virus type 1 (HIV-1) gp120 CD4-binding site in a group of long term non progressor (LTNP) subjects.Retrovirology. 2009 Jan 15;6:4. doi: 10.1186/1742-4690-6-4. Retrovirology. 2009. PMID: 19146663 Free PMC article.
-
Dengue virus type 3 adaptive changes during epidemics in São Jose de Rio Preto, Brazil, 2006-2007.PLoS One. 2013 May 7;8(5):e63496. doi: 10.1371/journal.pone.0063496. Print 2013. PLoS One. 2013. PMID: 23667626 Free PMC article.
-
Wave-like spread of Ebola Zaire.PLoS Biol. 2005 Nov;3(11):e371. doi: 10.1371/journal.pbio.0030371. Epub 2005 Oct 25. PLoS Biol. 2005. PMID: 16231972 Free PMC article.
-
Adaptation of a diverse simian immunodeficiency virus population to a new host is revealed through a systematic approach to identify amino acid sites under selection.Mol Biol Evol. 2007 Mar;24(3):660-9. doi: 10.1093/molbev/msl194. Epub 2006 Dec 11. Mol Biol Evol. 2007. PMID: 17159231 Free PMC article.
References
-
- Bocharov, G., P. Klenerman, and S. Ehl. 2003. Modelling the dynamics of LCMV infection in mice. II. Compartmental structure and immunopathology. J. Theor. Biol. 221:349-378. - PubMed
-
- Booth, J. C., U. Kumar, D. Webster, J. Monjardino, and H. C. Thomas. 1998. Comparison of the rate of sequence variation in the hypervariable region of E2/NS1 region of hepatitis C virus in normal and hypogammaglobulinemic patients. Hepatology 27:223-227. - PubMed
-
- Cerino, A., M. Bissolati, A. Cividini, A. Nicosia, M. Esumi, N. Hayashi, K. Mizuno, R. Slobbe, P. Oudshoorn, E. Silini, M. Asti, and M. U. Mondelli. 1997. Antibody responses to the hepatitis C virus E2 protein: relationship to viraemia and prevalence in anti-HCV seronegative subjects. J. Med. Virol. 51:1-5. - PubMed
-
- Cohen, J. 1999. The scientific challenge of hepatitis C. Science 285:26-30. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

