High-throughput screening of the yeast kinome: identification of human serine/threonine protein kinases that phosphorylate the hepatitis C virus NS5A protein
- PMID: 15016873
- PMCID: PMC371080
- DOI: 10.1128/jvi.78.7.3502-3513.2004
High-throughput screening of the yeast kinome: identification of human serine/threonine protein kinases that phosphorylate the hepatitis C virus NS5A protein
Abstract
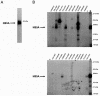
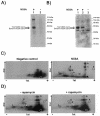
The hepatitis C virus NS5A protein plays a critical role in virus replication, conferring interferon resistance to the virus through perturbation of multiple intracellular signaling pathways. Since NS5A is a phosphoprotein, it is of considerable interest to understand the role of phosphorylation in NS5A function. In this report, we investigated the phosphorylation of NS5A by taking advantage of 119 glutathione S-transferase-tagged protein kinases purified from Saccharomyces cerevisiae to perform a global screening of yeast kinases capable of phosphorylating NS5A in vitro. A database BLAST search was subsequently performed by using the sequences of the yeast kinases that phosphorylated NS5A in order to identify human kinases with the highest sequence homologies. Subsequent in vitro kinase assays and phosphopeptide mapping studies confirmed that several of the homologous human protein kinases were capable of phosphorylating NS5A. In vivo phosphopeptide mapping revealed phosphopeptides common to those generated in vitro by AKT, p70S6K, MEK1, and MKK6, suggesting that these kinases may phosphorylate NS5A in mammalian cells. Significantly, rapamycin, an inhibitor commonly used to investigate the mTOR/p70S6K pathway, reduced the in vivo phosphorylation of specific NS5A phosphopeptides, strongly suggesting that p70S6 kinase and potentially related members of this group phosphorylate NS5A inside the cell. Curiously, certain of these kinases also play a major role in mRNA translation and antiapoptotic pathways, some of which are already known to be regulated by NS5A. The findings presented here demonstrate the use of high-throughput screening of the yeast kinome to facilitate the major task of identifying human NS5A protein kinases for further characterization of phosphorylation events in vivo. Our results suggest that this novel approach may be generally applicable to the screening of other protein biochemical activities by mechanistic class.
Figures

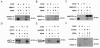


References
-
- Chan, T. O., S. E. Rittenhouse, and P. N. Tsichlis. 1999. AKT/PKB and other D3 phosphoinositide-regulated kinases: kinase activation by phosphoinositide-dependent phosphorylation. Annu. Rev. Biochem. 68:965-1014. - PubMed
-
- Deacon, K., and J. L. Blank. 1997. Characterization of the mitogen-activated protein kinase kinase 4 (MKK4)/c-Jun NH2-terminal kinase 1 and MKK3/p38 pathways regulated by MEK kinases 2 and 3. MEK kinase 3 activates MKK3 but does not cause activation of p38 kinase in vivo. J. Biol. Chem. 272:14489-14496. - PubMed
-
- Deacon, K., and J. L. Blank. 1999. MEK kinase 3 directly activates MKK6 and MKK7, specific activators of the p38 and c-Jun NH2-terminal kinases. J. Biol. Chem. 274:16604-16610. - PubMed
-
- Enomoto, N., I. Sakuma, Y. Asahina, M. Kurosaki, T. Murakami, C. Yamamoto, N. Izumi, F. Marumo, and C. Sato. 1995. Comparison of full-length sequences of interferon-sensitive and resistant hepatitis C virus 1b. Sensitivity to interferon is conferred by amino acid substitutions in the NS5A region. J. Clin. Investig. 96:224-230. - PMC - PubMed
-
- Enomoto, N., I. Sakuma, Y. Asahina, M. Kurosaki, T. Murakami, C. Yamamoto, Y. Ogura, N. Izumi, F. Marumo, and C. Sato. 1996. Mutations in the nonstructural protein 5A gene and response to interferon in patients with chronic hepatitis C virus 1b infection. N. Engl. J. Med. 334:77-81. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

