Effects of mutations of the initiation nucleotides on hepatitis C virus RNA replication in the cell
- PMID: 15016884
- PMCID: PMC371060
- DOI: 10.1128/jvi.78.7.3633-3643.2004
Effects of mutations of the initiation nucleotides on hepatitis C virus RNA replication in the cell
Abstract
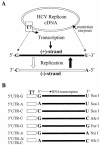
Replication of nearly all RNA viruses depends on a virus-encoded RNA-dependent RNA polymerase (RdRp). Our earlier work found that purified recombinant hepatitis C virus (HCV) RdRp (NS5B) was able to initiate RNA synthesis de novo by using purine (A and G) but not pyrimidine (C and U) nucleotides (G. Luo et al., J. Virol. 74:851-863, 2000). For most human RNA viruses, the initiation nucleotides of both positive- and negative-strand RNAs were found to be either an adenylate (A) or guanylate (G). To determine the nucleotide used for initiation and control of HCV RNA replication, a genetic mutagenesis analysis of the nucleotides at the very 5' and 3' ends of HCV RNAs was performed by using a cell-based HCV replicon replication system. Either a G or an A at the 5' end of HCV genomic RNA was able to efficiently induce cell colony formation, whereas a nucleotide C at the 5' end dramatically reduced the efficiency of cell colony formation. Likewise, the 3'-end nucleotide U-to-C mutation did not significantly affect the efficiency of cell colony formation. In contrast, a U-to-G mutation at the 3' end caused a remarkable decrease in cell colony formation, and a U-to-A mutation resulted in a complete abolition of cell colony formation. Sequence analysis of the HCV replicon RNAs recovered from G418-resistant Huh7 cells revealed several interesting findings. First, the 5'-end nucleotide G of the replicon RNA was changed to an A upon multiple rounds of replication. Second, the nucleotide A at the 5' end was stably maintained among all replicon RNAs isolated from Huh7 cells transfected with an RNA with a 5'-end A. Third, initiation of HCV RNA replication with a CTP resulted in a >10-fold reduction in the levels of HCV RNAs, suggesting that initiation of RNA replication with CTP was very inefficient. Fourth, the 3'-end nucleotide U-to-C and -G mutations were all reverted back to a wild-type nucleotide U. In addition, extra U and UU residues were identified at the 3' ends of revertants recovered from Huh7 cells transfected with an RNA with a nucleotide G at the 3' end. We also determined the 5'-end nucleotide of positive-strand RNA of some clinical HCV isolates. Either G or A was identified at the 5' end of HCV RNA genome depending on the specific HCV isolate. Collectively, these findings demonstrate that replication of positive-strand HCV RNA was preferentially initiated with purine nucleotides (ATP and GTP), whereas the negative-strand HCV RNA replication is invariably initiated with an ATP.
Figures

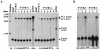

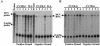



Similar articles
-
Mutagenesis analysis of the rGTP-specific binding site of hepatitis C virus RNA-dependent RNA polymerase.J Virol. 2005 Sep;79(18):11607-17. doi: 10.1128/JVI.79.18.11607-11617.2005. J Virol. 2005. PMID: 16140738 Free PMC article.
-
Selectable subgenomic and genome-length dicistronic RNAs derived from an infectious molecular clone of the HCV-N strain of hepatitis C virus replicate efficiently in cultured Huh7 cells.J Virol. 2002 Mar;76(6):2997-3006. doi: 10.1128/jvi.76.6.2997-3006.2002. J Virol. 2002. PMID: 11861865 Free PMC article.
-
Evidence for Internal Initiation of RNA Synthesis by the Hepatitis C Virus RNA-Dependent RNA Polymerase NS5B In Cellulo.J Virol. 2019 Sep 12;93(19):e00525-19. doi: 10.1128/JVI.00525-19. Print 2019 Oct 1. J Virol. 2019. PMID: 31315989 Free PMC article.
-
[Replication of hepatitis C virus genome].Uirusu. 2008 Dec;58(2):191-8. Uirusu. 2008. PMID: 19374197 Review. Japanese.
-
Mechanisms of viral mutation.Cell Mol Life Sci. 2016 Dec;73(23):4433-4448. doi: 10.1007/s00018-016-2299-6. Epub 2016 Jul 8. Cell Mol Life Sci. 2016. PMID: 27392606 Free PMC article. Review.
Cited by
-
Three-dimensional structure of the 3'X-tail of hepatitis C virus RNA in monomeric and dimeric states.RNA. 2017 Sep;23(9):1465-1476. doi: 10.1261/rna.060632.117. Epub 2017 Jun 19. RNA. 2017. PMID: 28630140 Free PMC article.
-
Full-length sequence analysis of hepatitis C virus genotype 3b strains and development of an in vivo infectious 3b cDNA clone.J Virol. 2023 Dec 21;97(12):e0092523. doi: 10.1128/jvi.00925-23. Epub 2023 Nov 21. J Virol. 2023. PMID: 38092564 Free PMC article.
-
Visualization and measurement of ATP levels in living cells replicating hepatitis C virus genome RNA.PLoS Pathog. 2012;8(3):e1002561. doi: 10.1371/journal.ppat.1002561. Epub 2012 Mar 1. PLoS Pathog. 2012. PMID: 22396648 Free PMC article.
-
Evidence that the polymerase of respiratory syncytial virus initiates RNA replication in a nontemplated fashion.Proc Natl Acad Sci U S A. 2010 Jun 1;107(22):10226-31. doi: 10.1073/pnas.0913065107. Epub 2010 May 17. Proc Natl Acad Sci U S A. 2010. PMID: 20479224 Free PMC article.
-
cis-Acting RNA elements in the hepatitis C virus RNA genome.Virus Res. 2015 Aug 3;206:90-8. doi: 10.1016/j.virusres.2014.12.029. Epub 2015 Jan 7. Virus Res. 2015. PMID: 25576644 Free PMC article. Review.
References
-
- Ackermann, M., and R. Padmanabhan. 2001. De novo synthesis of RNA by the dengue virus RNA-dependent RNA polymerase exhibits temperature dependence at the initiation but not elongation phase. J. Biol. Chem. 276:39926-39937. - PubMed
-
- Bartholomeusz, A., and P. Thompson. 1999. Flaviviridae polymerase and RNA replication. J. Viral Hepat. 6:261-270. - PubMed
-
- Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1975. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

