The subcellular distribution of multigene family 110 proteins of African swine fever virus is determined by differences in C-terminal KDEL endoplasmic reticulum retention motifs
- PMID: 15016891
- PMCID: PMC371041
- DOI: 10.1128/jvi.78.7.3710-3721.2004
The subcellular distribution of multigene family 110 proteins of African swine fever virus is determined by differences in C-terminal KDEL endoplasmic reticulum retention motifs
Abstract
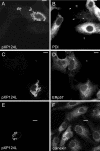
African swine fever virus (ASFV) is a large double-stranded DNA virus that replicates in discrete areas in the cytosol of infected cells called viral factories. Recent studies have shown that assembling virions acquire their internal envelopes through enwrapment by membranes derived from the endoplasmic reticulum (ER). However, the mechanisms that underlie the formation of viral factories and progenitor viral membranes are as yet unclear. Analysis of the published genome of the virus revealed a conserved multigene family that encodes proteins with hydrophobic signal sequences, indicating possible translocation into the ER lumen. Strikingly, two of these genes, XP124L and Y118L, encoded proteins with KDEL-like ER retention motifs. Analysis of XP124L and Y118L gene product by biochemical and immunofluorescence techniques showed that the proteins were localized to pre-Golgi compartments and that the KEDL motif at the C terminus of pXP124L was functional. XP124L expression, in the absence of other ASFV genes, had a dramatic effect on the contents of the ER that was dependent precisely on the C-terminal sequence KEDL. The normal subcellular distribution of a number of proteins resident to this important, cellular organelle was drastically altered in cells expressing wild-type XP124L gene product. PXP124L formed unusual perinuclear structures that contained resident ER proteins, as well as proteins of the ER-Golgi intermediate compartment. The data presented here hint at a role for MGF110 gene product in preparing the ER for its role in viral morphogenesis; this and other potential functions are discussed.
Figures



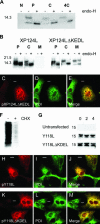

Similar articles
-
African swine fever virus is wrapped by the endoplasmic reticulum.J Virol. 1998 Mar;72(3):2373-87. doi: 10.1128/JVI.72.3.2373-2387.1998. J Virol. 1998. PMID: 9499098 Free PMC article.
-
Biochemical requirements of virus wrapping by the endoplasmic reticulum: involvement of ATP and endoplasmic reticulum calcium store during envelopment of African swine fever virus.J Virol. 2000 Mar;74(5):2151-60. doi: 10.1128/jvi.74.5.2151-2160.2000. J Virol. 2000. PMID: 10666244 Free PMC article.
-
Viroporin-like activity of the hairpin transmembrane domain of African swine fever virus B169L protein.J Virol. 2024 Aug 20;98(8):e0023124. doi: 10.1128/jvi.00231-24. Epub 2024 Jul 9. J Virol. 2024. PMID: 38980063 Free PMC article.
-
Retention and retrieval in the endoplasmic reticulum and the Golgi apparatus.Curr Opin Cell Biol. 1994 Aug;6(4):517-21. doi: 10.1016/0955-0674(94)90070-1. Curr Opin Cell Biol. 1994. PMID: 7986527 Free PMC article. Review.
-
Pathogenic Effects of Impaired Retrieval between the Endoplasmic Reticulum and Golgi Complex.Int J Mol Sci. 2019 Nov 9;20(22):5614. doi: 10.3390/ijms20225614. Int J Mol Sci. 2019. PMID: 31717602 Free PMC article. Review.
Cited by
-
African swine fever virus inhibits induction of the stress-induced proapoptotic transcription factor CHOP/GADD153.J Virol. 2004 Oct;78(19):10825-8. doi: 10.1128/JVI.78.19.10825-10828.2004. J Virol. 2004. PMID: 15367650 Free PMC article.
-
Comparative genomic and transcriptomic analyses of African swine fever virus strains.Comput Struct Biotechnol J. 2023 Aug 29;21:4322-4335. doi: 10.1016/j.csbj.2023.08.028. eCollection 2023. Comput Struct Biotechnol J. 2023. PMID: 37711186 Free PMC article.
-
Modulation of membrane traffic between endoplasmic reticulum, ERGIC and Golgi to generate compartments for the replication of bacteria and viruses.Semin Cell Dev Biol. 2009 Sep;20(7):828-33. doi: 10.1016/j.semcdb.2009.03.015. Semin Cell Dev Biol. 2009. PMID: 19508853 Free PMC article. Review.
-
Increased resolution of African swine fever virus genome patterns based on profile HMMs of protein domains.Virus Evol. 2020 Jun 19;6(2):veaa044. doi: 10.1093/ve/veaa044. eCollection 2020 Jul. Virus Evol. 2020. PMID: 32913663 Free PMC article.
-
The First African Swine Fever Viruses Detected in Wild Boar in Hong Kong, 2021-2023.Viruses. 2025 Jun 25;17(7):896. doi: 10.3390/v17070896. Viruses. 2025. PMID: 40733514 Free PMC article.
References
-
- Beinert, H., R. H. Holm, and E. Münck. 1997. Iron-sulfur clusters: nature's modular, multipurpose structures. Science 277:653-659. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

