RNA conformational classes
- PMID: 15016910
- PMCID: PMC390331
- DOI: 10.1093/nar/gkh333
RNA conformational classes
Abstract
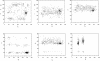
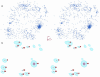
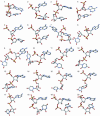
RNA exhibits a large diversity of conformations. Three thousand nucleotides of 23S and 5S ribosomal RNA from a structure of the large ribosomal subunit were analyzed in order to classify their conformations. Fourier averaging of the six 3D distributions of torsion angles and analyses of the resulting pseudo electron maps, followed by clustering of the preferred combinations of torsion angles were performed on this dataset. Eighteen non-A-type conformations and 14 A-RNA related conformations were discovered and their torsion angles were determined; their Cartesian coordinates are available.
Figures



References
-
- Ban N., Nissen,P., Hansen,J., Moore,P.B. and Steitz,T.A. (2000) The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science, 289, 905–920. - PubMed
-
- Tocilj A., Schlunzen,F., Janell,D., Gluhmann,M., Hansen,H.A.S., Harms,J., Bashan,A., Bartels,H., Agmon,I., Franceschi,F. et al. (1999) The small ribosomal subunit from Thermus thermophilus at 4.5 Å resolution: pattern fittings and the identification of a functional site. Proc. Natl Acad. Sci. USA, 96, 14252–14257. - PMC - PubMed
-
- Wimberly B.T., Brodersen,D.E., Clemons,W.M.,Jr, Morgan-Warren,R.J., Carter,A.P., Vonrhein,C., Hartsch,T. and Ramakrishnan,V.R. (2000) Structure of the 30S ribosomal subunit. Nature, 407, 327–332. - PubMed
-
- Yusupova G.Z., Yusupov,M.M., Cate,J.H.D. and Noller,H.F. (2001) The path of messenger RNA through the ribosome. Cell, 106, 233–241. - PubMed
-
- Kim S.-H., Berman,H.M., Seeman,N.C. and Newton,M.D. (1973) Seven basic conformations of nucleic acid structural units. Acta Crystallogr. B, 29, 703–710.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions

