Ubiquitin-mediated targeting of a mutant plasma membrane ATPase, Pma1-7, to the endosomal/vacuolar system in yeast
- PMID: 15020711
- PMCID: PMC404032
- DOI: 10.1091/mbc.e03-10-0727
Ubiquitin-mediated targeting of a mutant plasma membrane ATPase, Pma1-7, to the endosomal/vacuolar system in yeast
Abstract
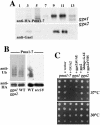
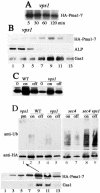
Pma1-7 is a mutant plasma membrane ATPase that is impaired in targeting to the cell surface at 37 degrees C and is delivered instead to the endosomal/vacuolar pathway for degradation. We have proposed that Pma1-7 is a substrate for a Golgibased quality control mechanism. By contrast with wild-type Pma1, Pma1-7 is ubiquitinated. Ubiquitination and endosomal targeting of Pma1-7 is dependent on the Rsp5-Bul1-Bul2 ubiquitin ligase protein complex but not the transmembrane ubiquitin ligase Tul1. Analysis of Pma1-7 ubiquitination in mutants blocked in protein transport at various steps of the secretory pathway suggests that ubiquitination occurs after ER exit but before endosomal entry. In the absence of ubiquitination in rsp5-1 cells, Pma1-7 is delivered to the cell surface and remains stable. Nevertheless, Pma1-7 remains impaired in association with detergent-insoluble glycolipid-enriched complexes in rsp5-1 cells, suggesting that ubiquitination is not the cause of Pma1-7 exclusion from rafts. In vps1 cells in which protein transport into the endosomal pathway is blocked, Pma1-7 is routed to the cell surface. On arrival at the plasma membrane in vps1 cells, Pma1-7 remains stable and its ubiquitination disappears, suggesting deubiquitination activity at the cell surface. We suggest that Pma1-7 sorting and fate are regulated by ubiquitination.
Figures
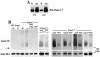


Similar articles
-
An endosome-to-plasma membrane pathway involved in trafficking of a mutant plasma membrane ATPase in yeast.Mol Biol Cell. 2000 Feb;11(2):579-92. doi: 10.1091/mbc.11.2.579. Mol Biol Cell. 2000. PMID: 10679016 Free PMC article.
-
Quality control of a mutant plasma membrane ATPase: ubiquitylation prevents cell-surface stability.J Cell Sci. 2006 Jan 15;119(Pt 2):360-9. doi: 10.1242/jcs.02749. J Cell Sci. 2006. PMID: 16410553
-
Loss of vacuolar H+-ATPase activity in organelles signals ubiquitination and endocytosis of the yeast plasma membrane proton pump Pma1p.J Biol Chem. 2014 Nov 14;289(46):32316-32326. doi: 10.1074/jbc.M114.574442. Epub 2014 Sep 30. J Biol Chem. 2014. PMID: 25271159 Free PMC article.
-
Ubiquitin and endocytic protein sorting.Essays Biochem. 2005;41:81-98. doi: 10.1042/EB0410081. Essays Biochem. 2005. PMID: 16250899 Review.
-
The ubiquitin code of yeast permease trafficking.Trends Cell Biol. 2010 Apr;20(4):196-204. doi: 10.1016/j.tcb.2010.01.004. Trends Cell Biol. 2010. PMID: 20138522 Review.
Cited by
-
Routing misfolded proteins through the multivesicular body (MVB) pathway protects against proteotoxicity.J Biol Chem. 2011 Aug 19;286(33):29376-29387. doi: 10.1074/jbc.M111.233346. Epub 2011 Jun 27. J Biol Chem. 2011. PMID: 21708947 Free PMC article.
-
Quality control: quality control at the plasma membrane: one mechanism does not fit all.J Cell Biol. 2014 Apr 14;205(1):11-20. doi: 10.1083/jcb.201310113. J Cell Biol. 2014. PMID: 24733583 Free PMC article. Review.
-
Specific α-arrestins negatively regulate Saccharomyces cerevisiae pheromone response by down-modulating the G-protein-coupled receptor Ste2.Mol Cell Biol. 2014 Jul;34(14):2660-81. doi: 10.1128/MCB.00230-14. Mol Cell Biol. 2014. PMID: 24820415 Free PMC article.
-
Rsp5 Ubiquitin ligase-mediated quality control system clears membrane proteins mistargeted to the vacuole membrane.J Cell Biol. 2019 Jan 7;218(1):234-250. doi: 10.1083/jcb.201806094. Epub 2018 Oct 25. J Cell Biol. 2019. PMID: 30361468 Free PMC article.
-
A mutant plasma membrane protein is stabilized upon loss of Yvh1, a novel ribosome assembly factor.Genetics. 2009 Mar;181(3):907-15. doi: 10.1534/genetics.108.100099. Epub 2008 Dec 29. Genetics. 2009. PMID: 19114459 Free PMC article.
References
-
- Armstrong, J., Patel, S., and Riddle, P. (1990). Lysosomal sorting mutants of coronavirus E1 protein, a Golgi membrane protein. J. Cell Sci. 95, 191–197. - PubMed
-
- Arvan, P., Zhao, X., Ramos-Castaneda, J., and Chang, A. (2002). Secretory pathway quality control in the Golgi, plasmalemmal, and endosomal systems. Traffic 3, 771-780. - PubMed
-
- Babst, M., Katzmann, D.J., Estepa-Sabal, E.J., Meerloo, T., and Emr, S.D. (2002). ESCRT-III: an endosome-associated heterooligomeric protein complex required for MVB sorting. Dev. Cell 3, 271–282. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

