Structural basis for HLA-DQ2-mediated presentation of gluten epitopes in celiac disease
- PMID: 15020763
- PMCID: PMC384714
- DOI: 10.1073/pnas.0306885101
Structural basis for HLA-DQ2-mediated presentation of gluten epitopes in celiac disease
Abstract
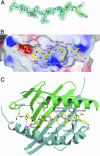
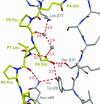
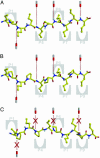
Celiac disease, also known as celiac sprue, is a gluten-induced autoimmune-like disorder of the small intestine, which is strongly associated with HLA-DQ2. The structure of DQ2 complexed with an immunogenic epitope from gluten, QLQPFPQPELPY, has been determined to 2.2-A resolution by x-ray crystallography. The glutamate at P6, which is formed by tissue transglutaminase-catalyzed deamidation, is an important anchor residue as it participates in an extensive hydrogen-bonding network involving Lys-beta71 of DQ2. The gluten peptide-DQ2 complex retains critical hydrogen bonds between the MHC and the peptide backbone despite the presence of many proline residues in the peptide that are unable to participate in amide-mediated hydrogen bonds. Positioning of proline residues such that they do not interfere with backbone hydrogen bonding results in a reduction in the number of registers available for gluten peptides to bind to MHC class II molecules and presumably impairs the likelihood of establishing favorable side-chain interactions. The HLA association in celiac disease can be explained by a superior ability of DQ2 to bind the biased repertoire of proline-rich gluten peptides that have survived gastrointestinal digestion and that have been deamidated by tissue transglutaminase. Finally, surface-exposed proline residues in the proteolytically resistant ligand were replaced with functionalized analogs, thereby providing a starting point for the design of orally active agents for blocking gluten-induced toxicity.
Figures
References
-
- Trier, J. S. (1991) N. Engl. J. Med. 325, 1709–1719. - PubMed
-
- Schmitz, J. (1992) in Coeliac Disease, ed. Marsh, M. N. (Blackwell, Oxford), pp. 17–48.
-
- Maki, M. & Collin, P. (1997) Lancet 349, 1755–1759. - PubMed
-
- Sollid, L. M. (2002) Nat. Rev. Immunol. 2, 647–655. - PubMed
-
- Sollid, L. M. & Thorsby, E. (1993) Gastroenterology 105, 910–922. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

