In vivo kinetics of Cajal body components
- PMID: 15024031
- PMCID: PMC1630494
- DOI: 10.1083/jcb.200311121
In vivo kinetics of Cajal body components
Abstract
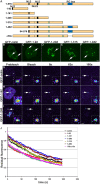
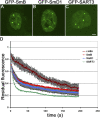
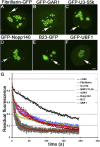
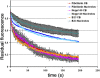
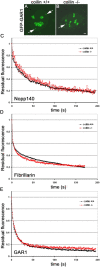
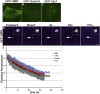
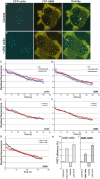
Cajal bodies (CBs) are subnuclear domains implicated in small nuclear ribonucleoprotein (snRNP) biogenesis. In most cell types, CBs coincide with nuclear gems, which contain the survival of motor neurons (SMN) complex, an essential snRNP assembly factor. Here, we analyze the exchange kinetics of multiple components of CBs and gems in living cells using photobleaching microscopy. We demonstrate differences in dissociation kinetics of CB constituents and relate them to their functions. Coilin and SMN complex members exhibit relatively long CB residence times, whereas components of snRNPs, small nucleolar RNPs, and factors shared with the nucleolus have significantly shorter residence times. Comparison of the dissociation kinetics of these shared proteins from either the nucleolus or the CB suggests the existence of compartment-specific retention mechanisms. The dynamic properties of several CB components do not depend on their interaction with coilin because their dissociation kinetics are unaltered in residual nuclear bodies of coilin knockout cells. Photobleaching and fluorescence resonance energy transfer experiments demonstrate that coilin and SMN can interact within CBs, but their interaction is not the major determinant of their residence times. These results suggest that CBs and gems are kinetically independent structures.
Figures