Palm mutants in DNA polymerases alpha and eta alter DNA replication fidelity and translesion activity
- PMID: 15024063
- PMCID: PMC371111
- DOI: 10.1128/MCB.24.7.2734-2746.2004
Palm mutants in DNA polymerases alpha and eta alter DNA replication fidelity and translesion activity
Abstract
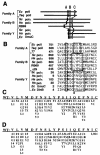
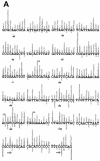
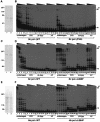
We isolated active mutants in Saccharomyces cerevisiae DNA polymerase alpha that were associated with a defect in error discrimination. Among them, L868F DNA polymerase alpha has a spontaneous error frequency of 3 in 100 nucleotides and 570-fold lower replication fidelity than wild-type (WT) polymerase alpha. In vivo, mutant DNA polymerases confer a mutator phenotype and are synergistic with msh2 or msh6, suggesting that DNA polymerase alpha-dependent replication errors are recognized and repaired by mismatch repair. In vitro, L868F DNA polymerase alpha catalyzes efficient bypass of a cis-syn cyclobutane pyrimidine dimer, extending the 3' T 26000-fold more efficiently than the WT. Phe34 is equivalent to residue Leu868 in translesion DNA polymerase eta, and the F34L mutant of S. cerevisiae DNA polymerase eta has reduced translesion DNA synthesis activity in vitro. These data suggest that high-fidelity DNA synthesis by DNA polymerase alpha is required for genomic stability in yeast. The data also suggest that the phenylalanine and leucine residues in translesion and replicative DNA polymerases, respectively, might have played a role in the functional evolution of these enzyme classes.
Figures


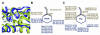
References
-
- Blasco, M. A., J. M. Lazaro, L. Blanco, and M. Salas. 1993. Phi 29 DNA polymerase active site. The conserved amino acid motif “Kx3NSxYG” is involved in template-primer binding and dNTP selection. J. Biol. Chem. 268:16763-16770. - PubMed
-
- Boudsocq, F., H. Ling, W. Yang, and R. Woodgate. 2002. Structure-based interpretation of missense mutations in Y-family DNA polymerases and their implications for polymerase function and lesion bypass. DNA Repair (Amsterdam) 1:343-358. - PubMed
-
- Capizzi, R. L., and J. W. Jameson. 1973. A table for the estimation of the spontaneous mutation rate of cells in culture. Mutat. Res. 17:147-148. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
