The essential WD repeat protein Swd2 has dual functions in RNA polymerase II transcription termination and lysine 4 methylation of histone H3
- PMID: 15024081
- PMCID: PMC371121
- DOI: 10.1128/MCB.24.7.2932-2943.2004
The essential WD repeat protein Swd2 has dual functions in RNA polymerase II transcription termination and lysine 4 methylation of histone H3
Abstract
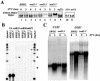
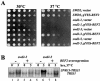
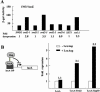
Swd2, an essential WD repeat protein in Saccharomyces cerevisiae, is a component of two very different complexes: the cleavage and polyadenylation factor CPF and the Set1 methylase, which modifies lysine 4 of histone H3 (H3-K4). It was not known if Swd2 is important for the function of either of these entities. We show here that, in extract from cells depleted of Swd2, cleavage and polyadenylation of the mRNA precursor in vitro are completely normal. However, temperature-sensitive mutations or depletion of Swd2 causes termination defects in some genes transcribed by RNA polymerase II. Overexpression of Ref2, a protein previously implicated in snoRNA 3' end formation and Swd2 recruitment to CPF, can rescue the growth and termination defects, indicating a functional interaction between the two proteins. Some swd2 mutations also significantly decrease global H3-K4 methylation and cause other phenotypes associated with loss of this chromatin modification, such as loss of telomere silencing, hydroxyurea sensitivity, and alterations in repression of INO1 transcription. Even though the two Swd2-containing complexes are both localized to actively transcribed genes, the allele specificities of swd2 defects suggest that the functions of Swd2 in mediating RNA polymerase II termination and H3-K4 methylation are not tightly coupled.
Figures






References
-
- Alen, C., N. A. Kent, H. S. Jones, J. O'Sullivan, A. Aranda, and N. J. Proudfoot. 2002. A role for chromatin remodeling in transcriptional termination by RNA polymerase II. Mol. Cell 10:1441-1452. - PubMed
-
- Betz, J. L., M. Chang, T. M. Washburn, S. E. Porter, C. L. Mueller, and J. A. Jaehning. 2002. Phenotypic analysis of Paf1/RNA polymerase II complex mutations reveals connections to cell cycle regulation, protein synthesis, and lipid and nucleic acid metabolism. Mol. Genet. Genomics 268:272-285. - PubMed
-
- Birse, C. E., L. Minvielle-Sebastia, B. A. Lee, W. Keller, and N. J. Proudfoot. 1998. Coupling termination of transcription to messenger RNA maturation in yeast. Science 280:298-301. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
