Small interfering RNA production by enzymatic engineering of DNA (SPEED)
- PMID: 15024103
- PMCID: PMC397411
- DOI: 10.1073/pnas.0400551101
Small interfering RNA production by enzymatic engineering of DNA (SPEED)
Abstract
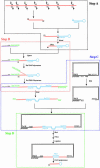
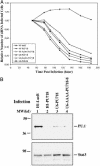
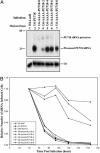
Small interfering RNAs (siRNAs) potently silence expression of target genes. In principle siRNA libraries can be used to perform effective genome-scale functional genetic screens in mammalian cells, but their development has been hampered by the need to chemically synthesize thousands of oligonucleotides and to incorporate them into expression vectors. We have developed a technology to efficiently convert a double-stranded cDNA library into a retroviral siRNA library in which 21-base siRNAs are produced in infected cells at high levels and efficiently block expression of their target genes. The key steps are the generation of random cDNA fragments that are fused to a hairpin linker, cleavage with the MmeI endonuclease that creates 20- to 21-bp cDNA fragments, conversion to a double-stranded DNA that contains two copies of the cDNA insert in a head-to-head palindrome, and insertion of the construct downstream of a polymerase III promoter. We constructed a siRNA library with 3 x 10(6) clones from a mouse embryo cDNA library; siRNAs were found against many different genes; and multiple siRNAs can be generated from a single mRNA. We further showed that specific siRNAs were efficiently produced in stably infected mammalian cells and resulted in significant and specific reduction of their target mRNAs. Because no prior knowledge about target transcripts is needed, a cDNA-derived siRNA library will generate siRNAs against unknown transcripts and genes. Finally, cDNA-derived siRNA libraries can be readily generated from any cell type or species, enabling genome-wide functional screens in many biological systems.
Figures


Comment in
-
Silence of the genes.Proc Natl Acad Sci U S A. 2004 Apr 13;101(15):5313-4. doi: 10.1073/pnas.0401209101. Epub 2004 Apr 5. Proc Natl Acad Sci U S A. 2004. PMID: 15067116 Free PMC article. Review. No abstract available.
Similar articles
-
Alternative approach to generate shRNA from cDNA.Biotechniques. 2005 Apr;38(4):629-32. doi: 10.2144/05384RN02. Biotechniques. 2005. PMID: 15884681
-
PCR-based generation of shRNA libraries from cDNAs.BMC Biotechnol. 2006 Jun 21;6:28. doi: 10.1186/1472-6750-6-28. BMC Biotechnol. 2006. PMID: 16790063 Free PMC article.
-
A plasmid-based system for expressing small interfering RNA libraries in mammalian cells.BMC Cell Biol. 2004 Apr 30;5:16. doi: 10.1186/1471-2121-5-16. BMC Cell Biol. 2004. PMID: 15119963 Free PMC article.
-
Genome-wide application of RNAi to the discovery of potential drug targets.FEBS Lett. 2005 Oct 31;579(26):5988-95. doi: 10.1016/j.febslet.2005.08.015. Epub 2005 Aug 22. FEBS Lett. 2005. PMID: 16153642 Review.
-
Approaches for chemically synthesized siRNA and vector-mediated RNAi.FEBS Lett. 2005 Oct 31;579(26):5974-81. doi: 10.1016/j.febslet.2005.08.070. Epub 2005 Sep 20. FEBS Lett. 2005. PMID: 16199038 Review.
Cited by
-
Function-based gene identification using enzymatically generated normalized shRNA library and massive parallel sequencing.Proc Natl Acad Sci U S A. 2010 Apr 20;107(16):7377-82. doi: 10.1073/pnas.1003055107. Epub 2010 Apr 5. Proc Natl Acad Sci U S A. 2010. PMID: 20368428 Free PMC article.
-
A novel method to convert a DNA fragment inserted into a plasmid to an inverted repeat structure.Mol Biotechnol. 2012 Jan;50(1):18-27. doi: 10.1007/s12033-011-9408-4. Mol Biotechnol. 2012. PMID: 21516519
-
Vector-based siRNA delivery strategies for high-throughput screening of novel target genes.J RNAi Gene Silencing. 2005 Jul 27;1(1):5-11. J RNAi Gene Silencing. 2005. PMID: 19771198 Free PMC article.
-
Strand antagonism in RNAi: an explanation of differences in potency between intracellularly expressed siRNA and shRNA.Nucleic Acids Res. 2012 Feb;40(4):1797-806. doi: 10.1093/nar/gkr927. Epub 2011 Oct 28. Nucleic Acids Res. 2012. PMID: 22039150 Free PMC article.
-
Antiviral RNAi: translating science towards therapeutic success.Pharm Res. 2011 Dec;28(12):2966-82. doi: 10.1007/s11095-011-0549-8. Epub 2011 Aug 9. Pharm Res. 2011. PMID: 21826573 Free PMC article. Review.
References
-
- Fire, A., Xu, S. Q., Montgomery, M. K., Kostas, S. A., Driver, S. E. & Mello, C. C. (1998) Nature 391, 806–811. - PubMed
-
- Hannon, G. J. (2002) Nature 418, 244–251. - PubMed
-
- McManus, M. T. & Sharp, P. A. (2002) Nat. Rev. Genet. 3, 737–747. - PubMed
-
- Paddison, P. J. & Hannon, G. J. (2003) Curr. Opin. Mol. Ther. 5, 217–224. - PubMed
-
- Ashrafi, K., Chang, F. Y., Watts, J. L., Fraser, A. G., Kamath, R. S., Ahringer, J. & Ruvkun, G. (2003) Nature 421, 268–272. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

