Coordinate enhancers share common organizational features in the Drosophila genome
- PMID: 15026577
- PMCID: PMC374333
- DOI: 10.1073/pnas.0400611101
Coordinate enhancers share common organizational features in the Drosophila genome
Abstract
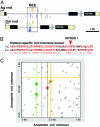
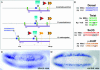
The evolution of animal diversity depends on changes in the regulation of a relatively fixed set of protein-coding genes. To understand how these changes might arise, we examined the organization of shared sequence motifs in four coordinately regulated neurogenic enhancers that direct similar patterns of gene expression in the early Drosophila embryo. All four enhancers possess similar arrangements of a subset of putative regulatory elements. These shared features were used to identify a neurogenic enhancer in the distantly related Anopheles genome. We suggest that the constrained organization of metazoan enhancers may be essential for their ability to produce precise patterns of gene expression during development. Organized binding sites should facilitate the identification of regulatory codes that link primary DNA sequence information with predicted patterns of gene activity.
Figures


References
-
- Thanos, D. & Maniatis, T. (1995) Cell 83, 1091-1100. - PubMed
-
- Belvin, M. P. & Anderson, K. V. (1996) Annu. Rev. Cell Dev. Biol. 12, 393-416. - PubMed
-
- Rusch, J. & Levine, M. (1996) Curr. Opin. Genet. Dev. 6, 416-423. - PubMed
-
- Drier, E. A. & Steward, R. (1997) Semin. Cancer Biol. 8, 83-92. - PubMed
-
- Stathopoulos, A. & Levine, M. (2002) Dev. Biol. 246, 57-67. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

