Human glutaminyl cyclase and bacterial zinc aminopeptidase share a common fold and active site
- PMID: 15028118
- PMCID: PMC368447
- DOI: 10.1186/1741-7007-2-2
Human glutaminyl cyclase and bacterial zinc aminopeptidase share a common fold and active site
Abstract
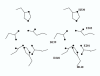
Background: Glutaminyl cyclase (QC) forms the pyroglutamyl residue at the amino terminus of numerous secretory peptides and proteins. We previously proposed the mammalian QC has some features in common with zinc aminopeptidases. We now have generated a structural model for human QC based on the aminopeptidase fold (pdb code 1AMP) and mutated the apparent active site residues to assess their role in QC catalysis.
Results: The structural model proposed here for human QC, deposited in the protein databank as 1MOI, is supported by a variety of fold prediction programs, by the circular dichroism spectrum, and by the presence of the disulfide. Mutagenesis of the six active site residues present in both 1AMP and QC reveal essential roles for the two histidines (140 and 330, QC numbering) and the two glutamates (201 and 202), while the two aspartates (159 and 248) appear to play no catalytic role. ICP-MS analysis shows less than stoichiometric zinc (0.3:1) in the purified enzyme.
Conclusions: We conclude that human pituitary glutaminyl cyclase and bacterial zinc aminopeptidase share a common fold and active site residues. In contrast to the aminopeptidase, however, QC does not appear to require zinc for enzymatic activity.
Figures


References
-
- Messer M, Ottesen M. Isolation and properties of glutamine cyclotransferase of dried papaya latex. C R Trav Lab Carlsberg. 1965;35:1–24. - PubMed
-
- Busby WH, Jr, Quackenbush GE, Humm J, Youngblood WW, Kizer JS. An enzyme that converts glutaminyl-peptides into pyroglutamyl-peptides. J Biol Chem. 1987;262:8532–8536. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

