Transcriptional analysis of spo0A overexpression in Clostridium acetobutylicum and its effect on the cell's response to butanol stress
- PMID: 15028679
- PMCID: PMC374416
- DOI: 10.1128/JB.186.7.1959-1971.2004
Transcriptional analysis of spo0A overexpression in Clostridium acetobutylicum and its effect on the cell's response to butanol stress
Abstract
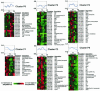
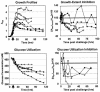
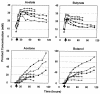
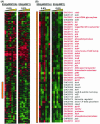
Spo0A is the regulator of stationary-phase events and is required for transcription of solvent formation genes in Clostridium acetobutylicum. In order to elucidate the role of spo0A in differentiation, we performed transcriptional analysis of 824(pMSPOA) (a spo0A-overexpressing C. acetobutylicum strain with enhanced sporulation) against a plasmid control strain. DNA microarray data were contrasted to data from a spo0A knockout strain (SKO1) that neither sporulates nor produces solvents. Transcripts of fatty acid metabolism genes, motility and chemotaxis genes, heat shock protein genes, and genes encoding the Fts family of cell division proteins were differentially expressed in the two strains, suggesting that these genes play roles in sporulation and the solvent stress response. 824(pMSPOA) alone showed significant downregulation of many glycolytic genes in stationary phase, which is consistent with metabolic flux analysis data. Surprisingly, spo0A overexpression resulted in only nominal transcriptional changes of regulatory genes (abrB and sigF) whose expression was significantly altered in SKO1. Overexpression of spo0A imparted increased tolerance and prolonged metabolism in response to butanol stress. While most of the differentially expressed genes appear to be part of a general stress response (similar to patterns in two plasmid control strains and a groESL-overexpressing strain), several genes were expressed at higher levels at early time points after butanol challenge only in 824(pMSPOA). Most of these genes were related to butyryl coenzyme A and butyrate formation and/or assimilation, but they also included the cell division gene ftsX, the gyrase subunit-encoding genes gyrB and gyrA, DNA synthesis and repair genes, and fatty acid synthesis genes, all of which might play a role in the immediate butanol stress response, and thus in enhanced butanol tolerance.
Figures






References
-
- Buday, Z., J. C. Linden, and M. N. Karim. 1990. Improved acetone butanol fermentation analysis using subambient HPLC column temperature. Enzyme Microb. Technol. 12:24-27.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

