Restricted distribution of the butyrate kinase pathway among butyrate-producing bacteria from the human colon
- PMID: 15028695
- PMCID: PMC374397
- DOI: 10.1128/JB.186.7.2099-2106.2004
Restricted distribution of the butyrate kinase pathway among butyrate-producing bacteria from the human colon
Abstract
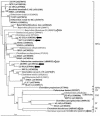
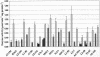
The final steps in butyrate synthesis by anaerobic bacteria can occur via butyrate kinase and phosphotransbutyrylase or via butyryl-coenzyme A (CoA):acetate CoA-transferase. Degenerate PCR and enzymatic assays were used to assess the presence of butyrate kinase among 38 anaerobic butyrate-producing bacterial isolates from human feces that represent three different clostridial clusters (IV, XIVa, and XVI). Only four strains were found to possess detectable butyrate kinase activity. These were also the only strains to give PCR products (verifiable by sequencing) with degenerate primer pairs designed within the butyrate kinase gene or between the linked butyrate kinase/phosphotransbutyrylase genes. Further analysis of the butyrate kinase/phosphotransbutyrylase genes of one isolate, L2-50, revealed similar organization to that described previously from different groups of clostridia, along with differences in flanking sequences and phylogenetic relationships. Butyryl-CoA:acetate CoA-transferase activity was detected in all 38 strains examined, suggesting that it, rather than butyrate kinase, provides the dominant route for butyrate formation in the human colonic ecosystem that contains a constantly high concentration of acetate.
Figures


References
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
-
- Atlas, R. M. 1997. In L. C. Parks (ed.), Handbook of microbiological media, p. 1245. CRC Press, Cleveland, Ohio.
-
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1998. Current protocols in molecular biology. John Wiley & Sons Inc., New York, N.Y.
-
- Barcenilla, A. 1999. Ph.D. thesis. Robert Gordon University, Aberdeen, United Kingdom.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

