Bacillus anthracis and Bacillus cereus PcrA helicases can support DNA unwinding and in vitro rolling-circle replication of plasmid pT181 of Staphylococcus aureus
- PMID: 15028705
- PMCID: PMC374418
- DOI: 10.1128/JB.186.7.2195-2199.2004
Bacillus anthracis and Bacillus cereus PcrA helicases can support DNA unwinding and in vitro rolling-circle replication of plasmid pT181 of Staphylococcus aureus
Abstract
Replication of rolling-circle replicating (RCR) plasmids in gram-positive bacteria requires the unwinding of initiator protein-nicked plasmid DNA by the PcrA helicase. In this report, we demonstrate that heterologous PcrA helicases from Bacillus anthracis and Bacillus cereus are capable of unwinding Staphylococcus aureus plasmid pT181 from the initiator-generated nick and promoting in vitro replication of the plasmid. These helicases also physically interact with the RepC initiator protein of pT181. The ability of PcrA helicases to unwind noncognate RCR plasmids may contribute to the broad-host-range replication and dissemination of RCR plasmids in gram-positive bacteria.
Figures
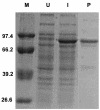

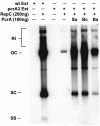


References
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
-
- Bird, L. E., S. Subramanya, and D. B. Wigley. 1998. Helicases: a unifying structural theme? Curr. Opin. Struct. Biol. 8:14-18. - PubMed
-
- Bruand, C., and S. D. Ehrlich. 2000. UvrD-dependent replication of rolling-circle plasmids in Escherichia coli. Mol. Microbiol. 35:204-210. - PubMed
-
- Chang, T.-L., M. G. Kramer, R. A. Ansari, and S. A. Khan. 2000. Role of individual monomers of a dimeric initiator protein in the initiation and termination of plasmid rolling circle replication. J. Biol. Chem. 275:13529-13534. - PubMed
-
- Chang, T.-L., A. Naqvi, S. P. Anand, M. G. Kramer, R. Munshi, and S. A. Khan. 2002. Biochemical characterization of the Staphylococcus aureus PcrA helicase and its role in plasmid rolling-circle replication. J. Biol. Chem. 277:45880-45886. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

