Blood mononuclear cell gene expression profiles characterize the oxidant, hemolytic, and inflammatory stress of sickle cell disease
- PMID: 15031206
- PMCID: PMC5560446
- DOI: 10.1182/blood-2003-08-2760
Blood mononuclear cell gene expression profiles characterize the oxidant, hemolytic, and inflammatory stress of sickle cell disease
Abstract
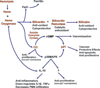
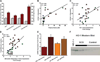
In sickle cell disease, deoxygenation of intra-erythrocytic hemoglobin S leads to hemoglobin polymerization, erythrocyte rigidity, hemolysis, and microvascular occlusion. Ischemia-reperfusion injury, plasma hemoglobin-mediated nitric oxide consumption, and free radical generation activate systemic inflammatory responses. To characterize the role of circulating leukocytes in sickle cell pathogenesis we performed global transcriptional analysis of blood mononuclear cells from 27 patients in steady-state sickle cell disease (10 patients treated and 17 patients untreated with hydroxyurea) compared with 13 control subjects. We used gender-specific gene expression to validate human microarray experiments. Patients with sickle cell disease demonstrated differential gene expression of 112 genes involved in heme metabolism, cell-cycle regulation, antioxidant and stress responses, inflammation, and angiogenesis. Inducible heme oxygenase-1 and downstream proteins biliverdin reductase and p21, a cyclin-dependent kinase, were up-regulated, potentially contributing to phenotypic heterogeneity and absence of atherosclerosis in patients with sickle cell disease despite endothelial dysfunction and vascular inflammation. Hydroxyurea therapy did not significantly affect leukocyte gene expression, suggesting that such therapy has limited direct anti-inflammatory activity beyond leukoreduction. Global transcriptional analysis of circulating leukocytes highlights the intense oxidant and inflammatory nature of steady-state sickle cell disease and provides insight into the broad compensatory responses to vascular injury.
Figures




References
-
- Bunn HF. Pathogenesis and treatment of sickle cell disease. N Engl J Med. 1997;337:762–769. - PubMed
-
- Noguchi CT, Haley JD, Abraham DJ, Schechter AN. Inhibition of sickle hemoglobin polymerization as a basis for therapeutic approaches to sickle cell anemia. In: Abraham DJ, editor. Burger’s Medicinal Chemistry. III. New York, NY: Wiley and Sons; 2003. pp. 443–479.
-
- Osarogiagbon UR, Choong S, Belcher JD, Vercellotti GM, Paller MS, Hebbel RP. Reperfusion injury pathophysiology in sickle transgenic mice. Blood. 2000;96:314–320. - PubMed
-
- Serjeant GR. Sickle Cell Disease. 2nd. New York, NY: Oxford University Press; 1992.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

