Arabidopsis peptide methionine sulfoxide reductase2 prevents cellular oxidative damage in long nights
- PMID: 15031406
- PMCID: PMC412865
- DOI: 10.1105/tpc.015818
Arabidopsis peptide methionine sulfoxide reductase2 prevents cellular oxidative damage in long nights
Abstract
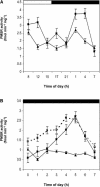
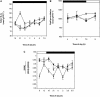
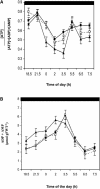
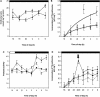
Peptide methionine sulfoxide reductase (PMSR) is a ubiquitous enzyme that repairs oxidatively damaged proteins. In Arabidopsis (Arabidopsis thaliana), a null mutation in PMSR2 (pmsr2-1), encoding a cytosolic isoform of the enzyme, exhibited reduced growth in short-day conditions. In wild-type plants, a diurnally regulated peak of total PMSR activity occurred at the end of the 16-h dark period that was absent in pmsr2-1 plants. This PMSR activity peak in the wild-type plant coincided with increased oxidative stress late in the dark period in the mutant. In pmsr2-1, the inability to repair proteins resulted in higher levels of their turnover, which in turn placed an increased burden on cellular metabolism. This caused increased respiration rates, leading to the observed higher levels of oxidative stress. In wild-type plants, the repair of damaged proteins by PMSR2 at the end of the night in a short-day diurnal cycle alleviates this potential burden on metabolism. Although PMSR2 is not absolutely required for viability of plants, the observation of increased damage to proteins in these long nights suggests the timing of expression of PMSR2 is an important adaptation for conservation of their resources.
Figures



References
-
- Affourtit, C., Krab, K., and Moore, A.L. (2001). Control of plant mitochondrial respiration. Biochim. Biophys. Acta 1504, 58–69. - PubMed
-
- Baker, N.R., Oxborough, K., Lawson, T., and Morisson, J.I. (2001). High resolution imaging of photosynthetic activities of tissues, cells and chloroplasts in leaves. J. Exp. Bot. 52, 615–621. - PubMed
-
- Berlett, B.S., and Stadtman, E.R. (1997). Protein oxidation in aging, disease, and oxidative stress. J. Biol. Chem. 372, 20313–20316. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

