A haplotype-specific Resistance gene regulated by BONZAI1 mediates temperature-dependent growth control in Arabidopsis
- PMID: 15031411
- PMCID: PMC412877
- DOI: 10.1105/tpc.020479
A haplotype-specific Resistance gene regulated by BONZAI1 mediates temperature-dependent growth control in Arabidopsis
Abstract
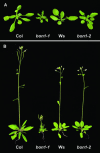
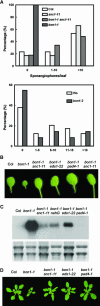
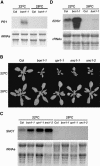
Plant growth homeostasis and defense responses are regulated by BONZAI1 (BON1), an evolutionarily conserved gene. Here, we show that growth regulation by BON1 is mediated through defense responses. BON1 is a negative regulator of a haplotype-specific Resistance (R) gene SNC1. The bon1-1 loss-of-function mutation activates SNC1, leading to constitutive defense responses and, consequently, reduced cell growth. In addition, a feedback amplification of the SNC1 gene involving salicylic acid is subject to temperature control, accounting for the regulation of growth and defense by temperature in bon1-1 and many other mutants. Thus, plant growth homeostasis involves the regulation of an R gene by BON1 and the intricate interplay between defense responses and temperature responses.
Figures


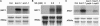

Similar articles
-
The TIR-NB-LRR gene SNC1 is regulated at the transcript level by multiple factors.Mol Plant Microbe Interact. 2007 Nov;20(11):1449-56. doi: 10.1094/MPMI-20-11-1449. Mol Plant Microbe Interact. 2007. PMID: 17977156
-
BON1 interacts with the protein kinases BIR1 and BAK1 in modulation of temperature-dependent plant growth and cell death in Arabidopsis.Plant J. 2011 Sep;67(6):1081-93. doi: 10.1111/j.1365-313X.2011.04659.x. Epub 2011 Jul 11. Plant J. 2011. PMID: 21623975
-
Plant growth homeostasis is controlled by the Arabidopsis BON1 and BAP1 genes.Genes Dev. 2001 Sep 1;15(17):2263-72. doi: 10.1101/gad.918101. Genes Dev. 2001. PMID: 11544183 Free PMC article.
-
Evidence that the BONZAI1/COPINE1 protein is a calcium- and pathogen-responsive defense suppressor.Plant Mol Biol. 2009 Jan;69(1-2):155-66. doi: 10.1007/s11103-008-9413-6. Epub 2008 Oct 15. Plant Mol Biol. 2009. PMID: 18855102
-
Complex regulation of an R gene SNC1 revealed by auto-immune mutants.Plant Signal Behav. 2012 Feb;7(2):213-6. doi: 10.4161/psb.18884. Epub 2012 Feb 1. Plant Signal Behav. 2012. PMID: 22415045 Free PMC article. Review.
Cited by
-
Constitutive Expresser of Pathogenesis Related Genes 1 Is Required for Pavement Cell Morphogenesis in Arabidopsis.PLoS One. 2015 Jul 20;10(7):e0133249. doi: 10.1371/journal.pone.0133249. eCollection 2015. PLoS One. 2015. PMID: 26193674 Free PMC article.
-
A complex network of additive and epistatic quantitative trait loci underlies natural variation of Arabidopsis thaliana quantitative disease resistance to Ralstonia solanacearum under heat stress.Mol Plant Pathol. 2020 Nov;21(11):1405-1420. doi: 10.1111/mpp.12964. Epub 2020 Sep 11. Mol Plant Pathol. 2020. PMID: 32914940 Free PMC article.
-
Modulation of ACD6 dependent hyperimmunity by natural alleles of an Arabidopsis thaliana NLR resistance gene.PLoS Genet. 2018 Sep 20;14(9):e1007628. doi: 10.1371/journal.pgen.1007628. eCollection 2018 Sep. PLoS Genet. 2018. PMID: 30235212 Free PMC article.
-
Comparative transcriptome analysis to reveal genes involved in wheat hybrid necrosis.Int J Mol Sci. 2014 Dec 16;15(12):23332-44. doi: 10.3390/ijms151223332. Int J Mol Sci. 2014. PMID: 25522166 Free PMC article.
-
Silencing of copine genes confers common wheat enhanced resistance to powdery mildew.Mol Plant Pathol. 2018 Jun;19(6):1343-1352. doi: 10.1111/mpp.12617. Epub 2017 Dec 26. Mol Plant Pathol. 2018. PMID: 28941084 Free PMC article.
References
-
- Austin, M.J., Muskett, P., Kahn, K., Feys, B.J., Jones, J.D., and Parker, J.E. (2002). Regulatory role of SGT1 in early R gene-mediated plant defenses. Science 295, 2077–2080. - PubMed
-
- Axtell, M.J., and Staskawicz, B.J. (2003). Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell 112, 369–377. - PubMed
-
- Azevedo, C., Sadanandom, A., Kitagawa, K., Freialdenhoven, A., Shirasu, K., and Schulze-Lefert, P. (2002). The RAR1 interactor SGT1, an essential component of R gene-triggered disease resistance. Science 295, 2073–2076. - PubMed
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

