The novel gene HOMOLOGOUS PAIRING ABERRATION IN RICE MEIOSIS1 of rice encodes a putative coiled-coil protein required for homologous chromosome pairing in meiosis
- PMID: 15031413
- PMCID: PMC412873
- DOI: 10.1105/tpc.020701
The novel gene HOMOLOGOUS PAIRING ABERRATION IN RICE MEIOSIS1 of rice encodes a putative coiled-coil protein required for homologous chromosome pairing in meiosis
Abstract
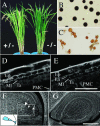
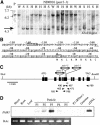
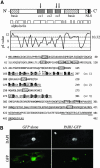
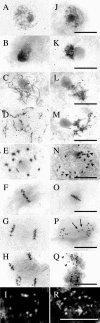
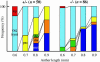
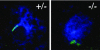
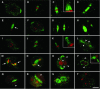
We have identified and characterized a novel gene, PAIR1 (HOMOLOGOUS PAIRING ABERRATION IN RICE MEIOSIS1), required for homologous chromosome pairing and cytokinesis in male and female meiocytes of rice (Oryza sativa). The pair1 mutation, tagged by the endogenous retrotransposon Tos17, exhibited meiosis-specific defects and resulted in complete sterility in male and female gametes. The PAIR1 gene encodes a 492-amino acid protein, which contains putative coiled-coil motifs in the middle, two basic regions at both termini, and a potential nuclear localization signal at the C terminus. Expression of the PAIR1 gene was detected in the early stages of flower development, in which the majority of the sporocytes had not entered meiosis. During prophase I of the pair1 meiocyte, all the chromosomes became entangled to form a compact sphere adhered to a nucleolus, and homologous pairing failed. At anaphase I and telophase I, chromosome nondisjunction and degenerated spindle formation resulted in multiple uneven spore production. However, chromosomal fragmentation frequent in plant meiotic mutants was never observed in all of the pair1 meiocytes. These observations clarify that the PAIR1 protein plays an essential role in establishment of homologous chromosome pairing in rice meiosis.
Figures

References
-
- Armstrong, S.J., Caryl, A.P., Jones, G.H., and Franklin, F.C. (2002). Asy1, a protein required for meiotic chromosome synapsis, localizes to axis-associated chromatin in Arabidopsis and Brassica. J. Cell Sci. 115, 3645–3655. - PubMed
-
- Armstrong, S.J., Franklin, F.C., and Jones, G.H. (2001). Nucleolus-associated telomere clustering and pairing precede meiotic chromosome synapsis in Arabidopsis thaliana. J. Cell Sci. 114, 4207–4217. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

