The population genetics and evolutionary epidemiology of RNA viruses
- PMID: 15031727
- PMCID: PMC7096949
- DOI: 10.1038/nrmicro863
The population genetics and evolutionary epidemiology of RNA viruses
Abstract
RNA viruses are ubiquitous intracellular parasites that are responsible for many emerging diseases, including AIDS and SARS. Here, we discuss the principal mechanisms of RNA virus evolution and highlight areas where future research is required. The rapidity of sequence change in RNA viruses means that they are useful experimental models for the study of evolution in general and it enables us to watch them change in 'real time', and retrace the spread through populations with molecular phylogenies. An understanding of the mechanisms of RNA virus sequence change is also crucial to predicting important aspects of their emergence and long-term evolution.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
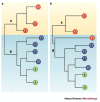
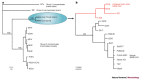
References
-
- Domingo E, Holland JJ. RNA virus mutations and fitness for survival. Ann. Rev. Microbiol. 1997;51:151–178. - PubMed
-
- Domingo E, Webster R, Holland J. Origin and Evolution of Viruses. 1999. - PubMed
-
- Grenfell BT, et al. Unifying the epidemiological and evolutionary dynamics of pathogens. Science. 2004;303:327–332. - PubMed
-
- Elena SF, Miralles R, Moya A. Frequency-dependent selection in a mammalian RNA virus. Evolution. 1997;5:984–987. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

