RalA activation at nascent lamellipodia of epidermal growth factor-stimulated Cos7 cells and migrating Madin-Darby canine kidney cells
- PMID: 15034142
- PMCID: PMC420081
- DOI: 10.1091/mbc.e03-11-0857
RalA activation at nascent lamellipodia of epidermal growth factor-stimulated Cos7 cells and migrating Madin-Darby canine kidney cells
Abstract
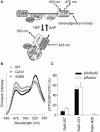
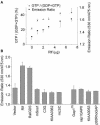
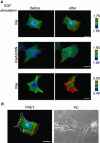
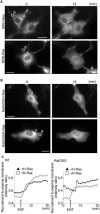
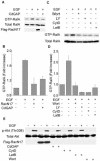
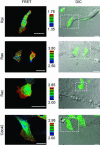
RalA, a member of the Ras-family GTPases, regulates various cellular functions such as filopodia formation, endocytosis, and exocytosis. On epidermal growth factor (EGF) stimulation, activated Ras recruits guanine nucleotide exchange factors (GEFs) for RalA, followed by RalA activation. By using fluorescence resonance energy transfer-based probes for RalA activity, we found that the EGF-induced RalA activation in Cos7 cells was restricted at the EGF-induced nascent lamellipodia, whereas under a similar condition both Ras activation and Ras-dependent translocation of Ral GEFs occurred more diffusely at the plasma membrane. This EGF-induced RalA activation was not observed when lamellipodial protrusion was suppressed by a dominant negative mutant of Rac1, a GTPase-activating protein for Cdc42, inhibitors of phosphatidylinositol 3-kinase, or inhibitors of actin polymerization. On the other hand, EGF-induced lamellipodial protrusion was inhibited by microinjection of the RalA-binding domains of RalBP1 and Sec5. Furthermore, we found that RalA activity was high at the lamellipodia of migrating Madin-Darby canine kidney cells and that the migration of Madin-Darby canine kidney cells was perturbed by the microinjection of RalBP1-RalA-binding domain. Thus, RalA activation is required for the induction of lamellipodia, and conversely, lamellipodial protrusion seems to be required for the RalA activation, suggesting the presence of a positive feedback loop between RalA activation and lamellipodial protrusion. Our observation also demonstrates that the spatial regulation of RalA is conducted by a mechanism distinct from the temporal regulation conducted by Ras-dependent plasma membrane recruitment of Ral guanine nucleotide exchange factors.
Figures


References
-
- Anderson, K.E., Coadwell, J., Stephens, L.R., and Hawkins, P.T. (1998). Translocation of PDK-1 to the plasma membrane is important in allowing PDK-1 to activate protein kinase B. Curr. Biol. 8, 684-691. - PubMed
-
- Aoki, K., Nakamura, T., and Matsuda, M. (2004). Spatio-temporal regulation of Rac1 and Cdc42 activity during nerve growth factor-induced neurite outgrowth in PC12 cells. J. Biol. Chem. 279, 713-719. - PubMed
-
- Bhattacharya, M., Anborgh, P.H., Babwah, A.V., Dale, L.B., Dobransky, T., Benovic, J.L., Feldman, R.D., Verdi, J.M., Rylett, R.J., and Ferguson, S.S. (2002). beta-Arrestins regulate a Ral-GDS Ral effector pathway that mediates cytoskeletal reorganization. Nat. Cell Biol. 4, 547-555. - PubMed
-
- Bhullar, R.P., and Seneviratne, H.D. (1996). Characterization of human platelet GTPase activating protein for the Ral GTP-binding protein. Biochim. Biophys. Acta 1311, 181-188. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

