Cloning, functional expression, and characterization of the raffinose oligosaccharide chain elongation enzyme, galactan:galactan galactosyltransferase, from common bugle leaves
- PMID: 15034167
- PMCID: PMC419815
- DOI: 10.1104/pp.103.036210
Cloning, functional expression, and characterization of the raffinose oligosaccharide chain elongation enzyme, galactan:galactan galactosyltransferase, from common bugle leaves
Abstract
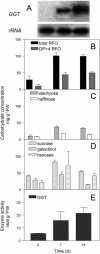
Galactan:galactan galactosyltransferase (GGT) is a unique enzyme of the raffinose family oligosaccharide (RFO) biosynthetic pathway. It catalyzes the chain elongation of RFOs without using galactinol (alpha-galactosyl-myoinositol) by simply transferring a terminal alpha-galactosyl residue from one RFO molecule to another one. Here, we report the cloning and functional expression of a cDNA encoding GGT from leaves of the common bugle (Ajuga reptans), a winter-hardy long-chain RFO-storing Lamiaceae. The cDNA comprises an open reading frame of 1215 bp. Expression in tobacco (Nicotiana plumbaginifolia) protoplasts resulted in a functional recombinant protein, which showed GGT activity like the previously described purified, native GGT enzyme. At the amino acid level, GGT shows high homologies (>60%) to acid plant alpha-galactosidases of the family 27 of glycosylhydrolases. It is clearly distinct from the family 36 of glycosylhydrolases, which harbor galactinol-dependent raffinose and stachyose synthases as well as alkaline alpha-galactosidases. Physiological studies on the role of GGT confirmed that GGT plays a key role in RFO chain elongation and carbon storage. When excised leaves were exposed to chilling temperatures, levels of GGT transcripts, enzyme activities, and long-chain RFO concentrations increased concomitantly. On a whole-plant level, chilling temperatures induced GGT expression mainly in the roots and fully developed leaves, both known RFO storage organs of the common bugle, indicating an adaptation of the metabolism from active growth to transient storage in the cold.
Figures



References
-
- Avigad G, Dey PM (1997) Carbohydrate metabolism: storage carbohydrates. In PM Dey, JB Harborne, eds, Plant Biochemistry. Academic Press, San Diego, pp 143–204
-
- Bachmann M, Inan C, Keller F (1995) Raffinose oligosaccharide storage. In MA Madore, WJ Lucas, eds, Carbon Partitioning and Source-Sink Interactions in Plants. American Society of Plant Physiologists, Rockville, MD, pp 215–225
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

