A post-genomic approach to understanding sphingolipid metabolism in Arabidopsis thaliana
- PMID: 15037448
- PMCID: PMC4242313
- DOI: 10.1093/aob/mch071
A post-genomic approach to understanding sphingolipid metabolism in Arabidopsis thaliana
Abstract
Aims: To highlight the importance of sphingolipids and their metabolites in plant biology.
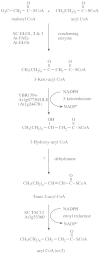
Scope: The completion of the arabidopsis genome provides a platform for the identification and functional characterization of genes involved in sphingolipid biosynthesis. Using the yeast Saccharomyces cerevisiae as an experimental model, this review annotates arabidopsis open reading frames likely to be involved in sphingolipid metabolism. A number of these open reading frames have already been subject to functional characterization, though the majority still awaits investigation. Plant-specific aspects of sphingolipid biology (such as enhanced long chain base heterogeneity) are considered in the context of the emerging roles for these lipids in plant form and function.
Conclusions: Arabidopsis provides an excellent genetic and post-genomic model for the characterization of the roles of sphingolipids in higher plants.
Figures
References
-
- BeaudoinF, Gable K, Sayanova O, Dunn T, Napier JA.2002. A Saccharomyces cerevisiae gene required for heterologous fatty acid elongase activity encodes a microsomal beta‐keto‐reductase. Journal of Biological Chemistry 277: 11481–11488. - PubMed
-
- BeckmannC, Rattke J, Sperling P, Heinz E, Boland W.2003. Stereochemistry of a bifunctional dihydroceramide delta 4‐desaturase/hydroxylase from Candida albicans; a key enzyme of sphingolipid metabolism. Organic and Biomolecular Chemistry 1: 2448–54. - PubMed
-
- BeelerT, Bacikova D, Gable K, Hopkins L, Johnson C, Slife H, Dunn T.1998. The Saccharomyces cerevisiae TSC10/YBR265w gene encoding 3‐ketosphinganine reductase is identified in a screen for temperature‐sensitive suppressors of the Ca2+‐sensitive csg2D mutant. Journal of Biological Chemistry 273: 30688–30694. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

