Structural basis of androgen receptor binding to selective androgen response elements
- PMID: 15037741
- PMCID: PMC387321
- DOI: 10.1073/pnas.0401123101
Structural basis of androgen receptor binding to selective androgen response elements
Abstract
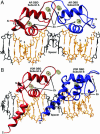
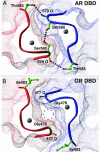
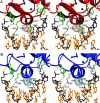
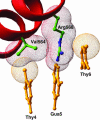
Steroid receptors bind as dimers to a degenerate set of response elements containing inverted repeats of a hexameric half-site separated by 3 bp of spacer (IR3). Naturally occurring selective androgen response elements have recently been identified that resemble direct repeats of the hexameric half-site (ADR3). The 3D crystal structure of the androgen receptor (AR) DNA-binding domain bound to a selective ADR3 reveals an unexpected head-to-head arrangement of the two protomers rather than the expected head-to-tail arrangement seen in nuclear receptors bound to response elements of similar geometry. Compared with the glucocorticoid receptor, the DNA-binding domain dimer interface of the AR has additional interactions that stabilize the AR dimer and increase the affinity for nonconsensus response elements. This increased interfacial stability compared with the other steroid receptors may account for the selective binding of AR to ADR3 response elements.
Figures


References
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

